Abstract
Lactobacillus sakei is a lactic acid bacterium important in food microbiology mainly due to its ability to ferment and preserve meat. The genome sequence of L. sakei strain 23K has revealed specialized metabolic capacities that reflect the bacterium’s adaption to meat products, and that differentiate it from other LAB. An extensive genomic diversity analysis was conducted to elucidate the core features of the species, and to provide a better comprehension of niche adaptation of the organism. Here, we describe the genomic comparison of 18 strains of L. sakei originating mainly from processed meat against the 23K strain by comparative genome hybridization. Pulsed field gel electrophoresis was used to estimate the genome sizes of the strains, which varied from 1.880 to 2.175 Mb, and the 23K genome was among the smallest. Consequently, a large part of the genome of this strain belongs to a common gene pool invariant in this species. The majority of genes important in adaption to meat products, the ability to flexibly use meat components, and robustness during meat processing and storage were conserved, such as genes involved in nucleoside scavenging, catabolism of arginine, and the ability to cope with changing redox and oxygen levels, which is indicative of the role these genes play in niche specialization within the L. sakei species. Moreover, an additional set of sequenced L. sakei genes beyond the 23K genome was present on the microarray used, and it was demonstrated that all the strains carry remnants of or complete bacteriocin operons. The genomic divergence corresponded mainly to five regions in the 23K genome, which showed features consistent with horizontal gene transfer. Carbohydrate-fermentation profiles of the strains were evaluated in light of the CGH data, and for most substrates, the genotypes were consistent with the phenotypes. We have demonstrated a highly conserved organization of the L. sakei genomes investigated, and the 23K strain is a suitable model organism to study core features of the L. sakei species.


Similar content being viewed by others
Abbreviations
- LAB:
-
Lactic acid bacteria
- CGH:
-
Comparative genome hybridization
- HGT:
-
Horizontal gene transfer
References
Aakra A, Nyquist OL, Snipen L, Reiersen TS, Nes IF (2007) Survey of genomic diversity among Enterococcus faecalis strains by microarray-based comparative genomic hybridization. Appl Environ Microbiol 73:2207–2217
Aasen IM, Moretro T, Katla T, Axelsson L, Storro I (2000) Influence of complex nutrients, temperature and pH on bacteriocin production by Lactobacillus sakei CCUG 42687. Appl Microbiol Biotechnol 53:159–166
Alpert CA, Crutz-Le Coq AM, Malleret C, Zagorec M (2003) Characterization of a theta-type plasmid from Lactobacillus sakei: a potential basis for low-copy-number vectors in lactobacilli. Appl Environ Microbiol 69:5574–5584
Axelsson L, Ahrné S (2000) Lactic acid bacteria. In: Priest FG, Goodfellow M (eds) Applied microbial systematics. Kluwer, Dordrechet, pp 365–386
Axelsson L, Holck A, Birkeland SE, Aukrust T, Blom H (1993) Cloning and nucleotide sequence of a gene from Lactobacillus sake Lb706 necessary for sakacin A production and immunity. Appl Environ Microbiol 59:2868–2875
Axelsson L, Katla T, Bjornslett M, Eijsink VG, Holck A (1998) A system for heterologous expression of bacteriocins in Lactobacillus sake. FEMS Microbiol Lett 168:137–143
Berthier F, Ehrlich SD (1999) Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int J Syst Bacteriol 49:997–1007
Bredholt S, Nesbakken T, Holck A (1999) Protective cultures inhibit growth of Listeria monocytogenes and Escherichia coli O157:H7 in cooked, sliced, vacuum- and gas-packaged meat. Int J Food Microbiol 53:43–52
Bredholt S, Nesbakken T, Holck A (2001) Industrial application of an antilisterial strain of Lactobacillus sakei as a protective culture and its effect on the sensory acceptability of cooked, sliced, vacuum-packaged meats. Int J Food Microbiol 66:191–196
Brussow H, Canchaya C, Hardt WD (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602
Bruyneel B, Vanderwoestyne M, Verstraete W (1989) Lactic acid bacteria—microorganisms able to grow in the absence of iron and copper. Biotechnol Lett 11:401–406
Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL (2009) Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol 2009:239–257
Canchaya C, Claesson MJ, Fitzgerald GF, van Sinderen D, O’Toole PW (2006) Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152:3185–3196
Chaillou S, Champomier-Verges MC, Cornet M, Crutz-Le Coq AM, Dudez AM, Martin V, Beaufils S, Darbon-Rongere E, Bossy R, Loux V, Zagorec M (2005) The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat Biotechnol 23:1527–1533
Chaillou S, Daty M, Baraige F, Dudez AM, Anglade P, Jones R, Alpert CA, Champomier-Verges MC, Zagorec M (2009) Intraspecies genomic diversity and natural population structure of the meat-borne lactic acid bacterium Lactobacillus sakei. Appl Environ Microbiol 75:970–980
Champomier Verges MC, Zuniga M, Morel-Deville F, Perez-Martinez G, Zagorec M, Ehrlich SD (1999) Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol Lett 180:297–304
Champomier-Verges MC, Chaillou S, Cornet M, Zagorec M (2001) Lactobacillus sakei: recent developments and future prospects. Res Microbiol 152:839–848
Champomier-Verges MC, Chaillou S, Cornet M, Zagorec M (2002) Erratum to “Lactobacillus sakei: recent developments and future prospects” [Research in Microbiology 152 (2001) 839]. Res Microbiol 153:115–123
Cheetham BF, Katz ME (1995) A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol 18:201–208
Chen YT, Liao TL, Wu KM, Lauderdale TL, Yan JJ, Huang IW, Lu MC, Lai YC, Liu YM, Shu HY, Wang JT, Su IJ, Tsai SF (2009) Genomic diversity of citrate fermentation in Klebsiella pneumoniae. BMC Microbiol 9:168
Chiaramonte F, Blugeon S, Chaillou S, Langella P, Zagorec M (2009) Behavior of the meat-borne bacterium Lactobacillus sakei during its transit through the gastrointestinal tracts of axenic and conventional mice. Appl Environ Microbiol 75:4498–4505
Chiaramonte F, Anglade P, Baraige F, Gratadoux JJ, Langella P, Champomier-Verges MC, Zagorec M (2010) Analysis of Lactobacillus sakei mutants selected after adaptation to the gastrointestinal tract of axenic mice. Appl Environ Microbiol 76:2932–2939
Claesson MJ, van Sinderen D, O’Toole PW (2007) The genus Lactobacillus—a genomic basis for understanding its diversity. FEMS Microbiol Lett 269:22–28
Condon S (1987) Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev 46:269–280
Cortez D, Forterre P, Gribaldo S (2009) A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol 10:R65
Dal Bello F, Walter J, Hammes WP, Hertel C (2003) Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb Ecol 45:455–463
Danielsen M (2002) Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98–103
Daubin V, Lerat E, Perriere G (2003) The source of laterally transferred genes in bacterial genomes. Genome Biol 4:R57
De Man JC, Rogosa M, Shape ME (1960) A medium for the cultivation of lactobacilli. J Appl Microbiol 23:130–135
Dong QJ, Wang Q, Xin YN, Li N, Xuan SY (2009) Comparative genomics of Helicobacter pylori. World J Gastroenterol 15:3984–3991
Dudez AM, Chaillou S, Hissler L, Stentz R, Champomier-Verges MC, Alpert CA, Zagorec M (2002) Physical and genetic map of the Lactobacillus sakei 23K chromosome. Microbiology 148:421–431
Duhutrel P, Bordat C, Wu TD, Zagorec M, Guerquin-Kern JL, Champomier-Verges MC (2010) Iron sources used by the nonpathogenic lactic acid bacterium Lactobacillus sakei as revealed by electron energy loss spectroscopy and secondary-ion mass spectrometry. Appl Environ Microbiol 76:560–565
Duwat P, Sourice S, Cesselin B, Lamberet G, Vido K, Gaudu P, Le Loir Y, Violet F, Loubiere P, Gruss A (2001) Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J Bacteriol 183:4509–4516
Eijsink VG, Brurberg MB, Middelhoven PH, Nes IF (1996) Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J Bacteriol 178:2232–2237
Garriga M, Hugas M, Aymerich T, Monfort JM (1993) Bacteriocinogenic activity of lactobacilli from fermented sausages. J Appl Bacteriol 75:142–148
Gevers D, Danielsen M, Huys G, Swings J (2003) Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl Environ Microbiol 69:1270–1275
Hagen BF, Naes H, Holck AL (2000) Meat starters have individual requirements for Mn2+. Meat Sci 55:161–168
Hallin PF, Binnewies TT, Ussery DW (2008) The genome BLASTatlas—a GeneWiz extension for visualization of whole-genome homology. Mol Biosyst 4:363–371
Hammes WP, Hertel C (1998) New developments in meat starter cultures. Meat Sci 49:125–138
Hammes WP, Vogel RF (1995) The genus Lactobacillus. In: Wood BJB, Hilzapfel WH (eds) The genera of lactic acid bacteria, vol 2. Blackie Academic, Professional, Glasgow, pp 19–54
Hertel C, Schmidt G, Fischer M, Oellers K, Hammes WP (1998) Oxygen-dependent regulation of the expression of the catalase gene katA of Lactobacillus sakei LTH677. Appl Environ Microbiol 64:1359–1365
Holck AL, Axelsson L, Huhne K, Krockel L (1994) Purification and cloning of sakacin 674, a bacteriocin from Lactobacillus sake Lb674. FEMS Microbiol Lett 115:143–149
Hugas M, Garriga M, Aymerich T, Monfort JM (1995) Inhibition of Listeria in dry fermented sausages by the bacteriocinogenic Lactobacillus sake CTC 494. J Appl Bacteriol 79:322–330
Jacquot R (1961) Organic constituents of fish and other aquatic animal foods. In: Borgstrom G (ed) Fish as food, vol 1. Academic, San Diego, pp 145–209
Katagiri H, Kitahara K, Fukami K (1934) The characteristics of the lactic acid bacteria isolated from moto, yeast mashes for sake manufacture. Part IV. Classification of the lactic acid bacteria. Bull Agric Chem Soc Jpn 10:156–157
Klaenhammer T, Altermann E, Arigoni F, Bolotin A, Breidt F, Broadbent J, Cano R, Chaillou S, Deutscher J, Gasson M, van de Guchte M, Guzzo J, Hartke A, Hawkins T, Hols P, Hutkins R, Kleerebezem M, Kok J, Kuipers O, Lubbers M, Maguin E, McKay L, Mills D, Nauta A, Overbeek R, Pel H, Pridmore D, Saier M, van Sinderen D, Sorokin A, Steele J, O’Sullivan D, de Vos W, Weimer B, Zagorec M, Siezen R (2002) Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82:29–58
Klein G, Dicks LMT, Pack A, Hack B, Zimmerman K, Dellaglio F, Reuter G (1996) Emended description of Lactobacillus sake (Katahiri, Katahara and Fukami) and Lactobacillus curvatus (Abo-Elnega Kandler): Numerical classification revealed by protein fingerprinting and identification based on biochemical patterns and DNA–DNA hybridizations. Int J Syst Bacteriol 46:367–376
Knauf HJ, Vogel RF, Hammes WP (1992) Cloning, sequence, and phenotypic expression of katA, which encodes the catalase of Lactobacillus sake LTH677. Appl Environ Microbiol 58:832–839
Koort J, Vandamme P, Schillinger U, Holzapfel W, Bjorkroth J (2004) Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int J Syst Evol Microbiol 54:1621–1626
Kralj S, van Geel-Schutten GH, Dondorff MM, Kirsanovs S, van der Maarel MJ, Dijkhuizen L (2004) Glucan synthesis in the genus Lactobacillus: isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 150:3681–3690
Lawrence JG, Ochman H (1997) Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol 44:383–397
Leistner L (2000) Basic aspects of food preservation by hurdle technology. Int J Food Microbiol 55:181–186
Leroi F, Joffraud JJ, Chevalier F, Cardinal M (1998) Study of the microbial ecology of cold-smoked salmon during storage at 8°C. Int J Food Microbiol 39:111–121
Leroy F, de Vuyst L (1999) The presence of salt and a curing agent reduces bacteriocin production by Lactobacillus sakei CTC 494, a potential starter culture for sausage fermentation. Appl Environ Microbiol 65:5350–5356
Leroy F, De Vuyst L (2000) Sakacins. In: Naidu AS (ed) Natural food antimicrobial systems. CRC Press, Boca Raton, pp 589–610
Lyhs U, Bjorkroth J, Korkeala H (1999) Characterisation of lactic acid bacteria from spoiled, vacuum-packaged, cold-smoked rainbow trout using ribotyping. Int J Food Microbiol 52:77–84
Magni C, de Mendoza D, Konings WN, Lolkema JS (1999) Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J Bacteriol 181:1451–1457
Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D (2006) Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA 103:15611–15616
Mathiesen G, Huehne K, Kroeckel L, Axelsson L, Eijsink VG (2005) Characterization of a new bacteriocin operon in sakacin P-producing Lactobacillus sakei, showing strong translational coupling between the bacteriocin and immunity genes. Appl Environ Microbiol 71:3565–3574
McLeod A, Nyquist OL, Snipen L, Naterstad K, Axelsson L (2008) Diversity of Lactobacillus sakei strains investigated by phenotypic and genotypic methods. Syst Appl Microbiol 31:393–403
McLeod A, Zagorec M, Champomier Verges MC, Naterstad K, Axelsson L (2010) Primary metabolism in Lactobacillus sakei by proteomic analysis. BMC Microbiol 10:120
Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M (2005) Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol 187:6119–6127
Moretro T, Naterstad K, Wang E, Aasen IM, Chaillou S, Zagorec M, Axelsson L (2005) Sakacin P non-producing Lactobacillus sakei strains contain homologues of the sakacin P gene cluster. Res Microbiol 156:949–960
Mortvedt CI, Nissen-Meyer J, Sletten K, Nes IF (1991) Purification and amino acid sequence of lactocin S, a bacteriocin produced by Lactobacillus sake L45. Appl Environ Microbiol 57:1829–1834
Murray AE, Lies D, Li G, Nealson K, Zhou J, Tiedje JM (2001) DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc Natl Acad Sci USA 98:9853–9858
Nigatu A (2000) Evaluation of numerical analyses of RAPD and API 50 CH patterns to differentiate Lactobacillus plantarum, L. fermentum, L. rhamnosus, L. sake, L. parabuchneri, L. gallinarum, L. casei, Weissella minor and related taxa isolated from kocho and tef. J Appl Microbiol 89:969–978
O’Sullivan O, O’Callaghan J, Sangrador-Vegas A, McAuliffe O, Slattery L, Kaleta P, Callanan M, Fitzgerald GF, Ross RP, Beresford T (2009) Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol 9:50
Obst M, Meding ER, Vogel RF, Hammes WP (1995) Two genes encoding the beta-galactosidase of Lactobacillus sake. Microbiology 141:3059–3066
Ouwehand AC, Salminen S, Isolauri E (2002) Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279–289
Pfeiler EA, Klaenhammer TR (2007) The genomics of lactic acid bacteria. Trends Microbiol 15:546–553
Riley MA, Wertz JE (2002) Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357–364
Rodriguez JM, Cintas LM, Casaus P, Suarez A, Hernandez PE (1995) PCR detection of the lactocin S structural gene in bacteriocin-producing lactobacilli from meat. Appl Environ Microbiol 61:2802–2805
Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A (2009) Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7:828–836
Schillinger U, Lucke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55:1901–1906
Schillinger U, Lücke F-K (1987) Identification of lactobacilli from meat and meat products. Food Microbiol 4:199–208
Siezen R, Boekhorst J, Muscariello L, Molenaar D, Renckens B, Kleerebezem M (2006) Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7:126
Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, van Hylckama Vlieg JE (2010) Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12:758–773
Simon L, Fremaux C, Cenatiempo Y, Berjeaud JM (2002) Sakacin g, a new type of antilisterial bacteriocin. Appl Environ Microbiol 68:6416–6420
Skaugen M, Abildgaard CI, Nes IF (1997) Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol Gen Genet 253:674–686
Skaugen M, Andersen EL, Christie VH, Nes IF (2002) Identification, characterization, and expression of a second, bicistronic, operon involved in the production of lactocin S in Lactobacillus sakei L45. Appl Environ Microbiol 68:720–727
Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31:265–273
Snipen L, Nyquist OL, Solheim M, Aakra A, Nes IF (2009) Improved analysis of bacterial CGH data beyond the log-ratio paradigm. BMC Bioinformatics 10:91
Solheim M, Aakra A, Snipen LG, Brede DA, Nes IF (2009) Comparative genomics of Enterococcus faecalis from healthy Norwegian infants. BMC Genomics 10:194
Stentz R, Zagorec M (1999) Ribose utilization in Lactobacillus sakei: analysis of the regulation of the rbs operon and putative involvement of a new transporter. J Mol Microbiol Biotechnol 1:165–173
Stentz R, Cornet M, Chaillou S, Zagorec M (2001) Adaption of Lactobacillus sakei to meat: a new regulatory mechanism of ribose utilization? INRA, EDP Sciences 81:131–138
Tichaczek PS, Vogel RF, Hammes WP (1994) Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH 673. Microbiology 140:361–367
Torriani S, Van Reenen GA, Klein G, Reuter G, Dellaglio F, Dicks LM (1996) Lactobacillus curvatus subsp. curvatus subsp. nov. and Lactobacillus curvatus subsp. melibiosus subsp. nov. and Lactobacillus sake subsp. sake subsp. nov. and Lactobacillus sake subsp. carnosus subsp. nov., new subspecies of Lactobacillus curvatus Abo-Elnaga and Kandler 1965 and Lactobacillus sake Katagiri, Kitahara, and Fukami 1934 (Klein et al. 1996, emended descriptions), respectively. Int J Syst Bacteriol 46:1158–1163
Vaughan A, Eijsink VG, Van Sinderen D (2003) Functional characterization of a composite bacteriocin locus from malt isolate Lactobacillus sakei 5. Appl Environ Microbiol 69:7194–7203
Vermeiren L, Devlieghere F, Debevere J (2004) Evaluation of meat born lactic acid bacteria as protective cultures for biopreservation of cooked meat products. Int J Food Microbiol 96:149–164
Vogel RF, Lohmann M, Nguyen M, Weller AN, Hammes WP (1993) Molecular characterization of Lactobacillus curvatus and L. sake isolated from sauerkraut and their application in sausage fermentations. J Appl Bacteriol 74:295–300
Weinberg ED (1997) The Lactobacillus anomaly: total iron abstinence. Perspect Biol Med 40:578–583
Zuniga M, Champomier-Verges M, Zagorec M, Perez-Martinez G (1998) Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol 180:4154–4159
Acknowledgments
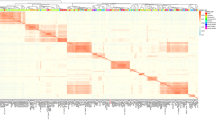
The authors would like to thank Professor Monique Zagorec, INRA, France, for the invitation to collaborate on production of the L. sakei microarray. We also thank Bjørn E. Kristiansen, the Norwegian Microarray Consortium (NMC), Oslo, for printing of the microarrays. We thank Dr. Dave Ussery, CBS, Denmark, for assistance given for the generation of Fig. 1.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by D. Ussery.
O. L. Nyquist and A. McLeod contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nyquist, O.L., McLeod, A., Brede, D.A. et al. Comparative genomics of Lactobacillus sakei with emphasis on strains from meat. Mol Genet Genomics 285, 297–311 (2011). https://doi.org/10.1007/s00438-011-0608-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-011-0608-1