Abstract
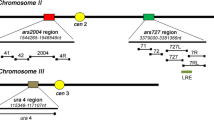
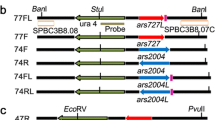
Eukaryotic chromosomal DNA replication is initiated by a highly conserved set of proteins that interact with cis-acting elements on chromosomes called replicators. Despite the conservation of replication initiation proteins, replicator sequences show little similarity from species to species in the small number of organisms that have been examined. Examination of replicators in other species is likely to reveal common features of replicators. We have examined a Kluyeromyces lactis replicator, KARS12, that functions as origin of DNA replication on plasmids and in the chromosome. It contains a 50-bp region with similarity to two other K. lactis replicators, KARS101 and the pKD1 replication origin. Replacement of the 50-bp sequence with an EcoRI site completely abrogated the ability of KARS12 to support plasmid and chromosomal DNA replication origin activity, demonstrating this sequence is a common feature of K. lactis replicators and is essential for function, possibly as the initiator protein binding site. Additional sequences up to 1 kb in length are required for efficient KARS12 function. Within these sequences are a binding site for a global regulator, Abf1p, and a region of bent DNA, both of which contribute to the activity of KARS12. These elements may facilitate protein binding, protein/protein interaction and/or nucleosome positioning as has been proposed for other eukaryotic origins of DNA replication.





Similar content being viewed by others
References
Aladjem MI, Fanning E (2004) The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5:686–691
Altman AL, Fanning E (2004) Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol Cell Biol 24:4138–4150
Ames JB, Hendricks KB, Strahl T, Huttner IG, Hamasaki N, Thorner J (2000) Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae. Biochem 40:12149–12161
Anachkova B, Djeliova V, Russev G (2005) Nuclear matrix support of DNA replication. J Cell Biochem 96:951–961
Austin RJ, Orr-Weaver TL, Bell SP (1999) Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev 13:2639–2649
Baker RT, Tobias JW, Varshavsky A (1992) Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem 267:23364–23375
Becker DM, Guarente L (1991) High-efficiency transformation of yeast by electroporation. Methods Enzymol 194:182–187
Bénard M, Lagnel C, Pallotta D, Pierron G (1996) Mapping of a replication origin within the promoter region of two unlinked, abundantly transcribed actin genes of Physarum polycephalum. Mol Cell Biol 16:968–976
Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475–477
Bielinsky AK, Blitzblau H, Beall EL, Ezrokhi M, Smith MS, Botchan MR, Gerbi SA (2001) Origin replication complex binding to a metazoan replication origin. Curr Biol 11:1427–1431
Boyer HW, Roulland-Dussoix D (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol 41:459–472
Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S cerevisiae. Cell 51:463–471
Chen XJ, Saliola M, Falcone C, Bianchi MM, Fukuhara H (1986) Sequence organization of the circular plasmid pKD1 from the yeast Kluyveromyces drosophilarum. Nucleic Acids Res 14:4471–4481
Cho G, Kim J, Rho HM, Jung G (1995) Structure–function analysis of the DNA binding domain of Saccharomyces cerevisiae ABF1. Nucleic Acid Res 23:2980–2987
Chuang RY, Kelly TJ (1999) The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA 96:2656–2661
Clyne RK, Kelly TJ (1995) Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J 14:6348–6357
Dai J, Chuang RY, Kelly TJ (2005) DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA 102:337–342
Das S, Hollenberg CP (1982) A high frequency transformation system for the yeast Kluyveromyces lactis. Curr Genet 6:123–128
Della Seta F, Ciafré SA, Marck C, Santoro B, Presutti C, Sentenac A, Bozzoni I (1990a) The ABF1 factor is the transcriptional activator of the L2 ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol 10:2437–2441
Della Seta F, Treich I, Buhler JM, Sentenac A (1990b) ABF1 binding sites in yeast RNA polymerase genes. J Biol Chem 265:15168–15175
DePamphilis ML (1999) Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21:5–16
Dijkwel PA, Vaughn JP, Hamlin JL (1991) Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol 11:3850–3859
Dubey DD, Kim SM, Todorov IT, Huberman JA (1996) Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol 6:467–473
Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I et al (2004) Genome evolution in yeasts. Nature 430:35–44
Fabiani L, Aragona M, Frontali L (1990) Isolation and sequence analysis of a K. lactis chromosomal element able to autonomously replicate in S. cerevisiae and K. lactis. Yeast 6:69–76
Fabiani L, Frontali L, Newlon CS (1996) Identification of an essential core element and stimulatory sequence in a Kluyveromyces lactis ARS element, KARS101. Mol Microbiol 19:757–766
Fabiani L, Irene C, Aragona M, Newlon CS (2001) A DNA replication origin and a replication fork barrier used in vivo in the circular plasmid pKD1. Mol Genet Genomics 266:326–335
Gavin KA, Hidaka M, Stillman B (1995) Conserved initiator proteins in eukaryotes. Science 270:1667–1671
Gerbi SA, Strezoska Z, Waggener JM (2003) Initiation of DNA replication in multicellular eukaryotes. J Struct Biol 140:17–30
Gonςalves PM, Maurer K, Mager WH, Planta RJ (1992) Kluyveromyces contains a functional ABF1-homologue. Nucleic Acids Res 20:2211–2215
Hoekstra R, Ferreira PM, Bootsma TC, Mager WH, Planta RJ (1992) Structure and expression of the ABF1-regulated ribosomal protein S33 gene in Kluyveromyces. Yeast 8:949–959
Hoekstra R, Groeneveld P, Van Verseveld HW, Stouthamer AH, Planta RJ (1994) Transcription regulation of ribosomal protein genes at different growth rates in continuous cultures of Kluyveromyces lactis. Yeast 10:637–651
Hurst ST, Rivier DH (1999) Identification of a compound origin of replication at the HMR-E locus in Saccharomyces cerevisiae. J Biol Chem 274:4155–4159
Irene C, Maciariello C, Cioci F, Camilloni G, Newlon CS, Fabiani L (2004) Identification of the sequences required for chromosomal replicator function in Kluyveromyces lactis. Mol Microbiol 51:1413–1423
Kim SM, Huberman JA (1998) Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol 18:7294–7303
Kim SM, Huberman JA (1999) Influence of a replication enhancer on the hierarchy of origin efficiencies within a cluster of DNA replication origins. J Mol Biol 288:867–882
Kong D, DePamphilis ML (2002) Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. EMBO J 21:5567–5576
Lin S, Kowalsky D (1997) Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol 17:5473–5484
Lipford JR, Bell SP (2001) Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7:21–30
Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817–823
Mead DA, Szczesna-Skorupa E, Kemper B (1986) Single-stranded DNA blue T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng 1:67–74
Miyake T, Loch CM, Li R (2002) Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol Cell Biol 22:505–516
Miyake T, Reese J, Loch CM, Auble DT, Li R (2004) Genome wide analysis of ARS (autonomous replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J Biol Chem 279:34865–34872
Mulder W, Scholten IH, Nagelkerken B, Grivell LA (1994) Isolation and characterization of the linked genes, FPS1 and QCR8, coding for farnesyl-diphosphate synthase and the 11 kDa subunit VIII of the mitochondrial bc1-complex in the yeast Kluyveromyces lactis. Biochim Biophis Acta 1219:713–718
Mulder W, Scholten IH, Grivell LA (1995) Distinct transcriptional regulation of a gene coding for a mitochondrial protein in the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis despite similar promoter structures. Mol Microbiol 17:813–824
Mysiak ME, Bleijenberg MH, Wyman C, Holthuizen PE, van der Vliet PC (2004) Bending of adenovirus origin DNA by Nuclear Factor I as shown by scanning force microscopy is required for optimal DNA replication. J Virol 78:1928–1935
Newlon CS (1996) DNA replication in yeast. In: De Pamphilis ML (ed) DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 873–914
Okuno Y, Satoh H, Sekiguchi M, Masukata H (1999) Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol Cell Biol 19:6699–6709
Polaczek P, Kwan K, Liberies DA, Campbell JL (1997) Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol Microbiol 26:261–275
Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B (2004) The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol 51:523–537
Rice P, Longden I, Bleasby A (2000) EMBOSS: The European molecular biology open software suite. Trends Genet 16:276–277
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA 74:54463–5467
Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication at A + T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4:1048–1053
Shpigelman ES, Trifonov EN, Bolshoy A (1993) CURVATURE: software for the analysis of curved DNA. Comput Appl Biosci 9:435–440
Takahashi T, Ohara E, Nishitani H, Masukata H (2003) Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. EMBO J 22:964–974
Theis JF, Newlon CS (2001) Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Mol Cell Biol 21:2790–2801
Venditti P, Costanzo G, Negri R, Camilloni G (1994) ABF1 contributes to the chromatin organization of Saccharomyces cerevisiae ARS1 B-domain. Biochim Biophis Acta 1219:677–689
Vernis L, Chasles M, Pasero P, Lepingle A, Gaillardin C, Fournier P (1999) Short DNA fragments without sequence similarity are sites for replication in the chromosome of the yeast Yarrowia lipolytica. Mol Biol Cell 10:757–769
Walker SS, Francesconi SC, Eisenberg S (1990) A DNA replication enhancer in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 87:4665–4669
Wang L, Lin CM, Brooks S, Cimbora D, Groudine M, Aladjem MI (2004) The human β-globin replication region consists of two modular independent replicators. Mol Cell Biol 24:3373–3386
Wilmes GM, Bell SP (2002) The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc Natl Acad Sci USA 99:101–106
Yarragudi A, Miyake T, Li R, Morse RH (2004) Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol 24:9152–9164
Acknowledgments
We thank Flavio della Seta for the affinity purified ABF1 fraction, Giorgio Camilloni, Silvia Ciafré and the members of the Newlon laboratory group for helpful discussions throughout the study. This work was supported by MURST (Ministero Università Ricerca Scientifica Tecnologica - Progetti Ateneo), Istituto Pasteur Fondazione Cenci Bolognetti and NIH (grant number GM35679 awarded to CSN). C.M. is a PhD student on Pasteurian Sciences, Università “La Sapienza”, Roma.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Irene, C., Maciariello, C., Micheli, G. et al. DNA elements modulating the KARS12 chromosomal replicator in Kluyveromyces lactis . Mol Genet Genomics 277, 287–299 (2007). https://doi.org/10.1007/s00438-006-0188-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0188-7