Abstract
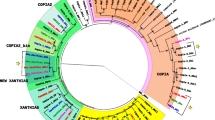
A new transposable element, Isis, is identified as a LTR retrotransposon in Drosophila buzzatii. DNA sequence analysis shows that Isis contains three long ORFs similar to gag, pol and env genes of retroviruses. The ORF1 exhibits sequence homology to matrix, capsid and nucleocapsid gag proteins and ORF2 encodes a putative protease (PR), a reverse transcriptase (RT), an Rnase H (RH) and an integrase (IN) region. The analysis of a putative env product, encoded by the env ORF3, shows a degenerated protein containing several stop codons. The molecular study of the putative proteins coded by this new element shows striking similarities to both Ulysses and Osvaldo elements, two LTR retrotransposons, present in D. virilis and D. buzzatii, respectively. Comparisons of the predicted Isis RT to several known retrotransposons show strong phylogenetic relationships to gypsy-like elements, particulary to Ulysses retrotransposon. Studies of Isis chromosomal distribution show a strong hybridization signal in centromeric and pericentromeric regions, and a scattered distribution along all chromosomal arms. The existence of insertional polymorphisms between different strains and high molecular weight bands by Southern blot suggests the existence of full-sized copies that have been active recently. The presence of euchromatic insertion sites coincident between Isis and Osvaldo could indicate preferential insertion sites of Osvaldo element into Isis sequence or vice versa. Moreover, the presence of Isis in different species of the buzzatii complex indicates the ancient origin of this element.













Similar content being viewed by others
References
Abe H, Kanehara M, Terada T, Ohbayashi F, Shimada T, Kawai S, Suzuki M, T Sugasaki T, Oshiki T (1998) Identification of novel random amplified polymorphic DNAs (RAPDs) on the W chromosome of the domesticated silkworm, Bombyx mori, and the wild silkworm B. mandarina, and their retrotransposable-related nucleotide sequences. Genes Genet Sys 73:243–254
Abe H, Ohbayashi F, Shimada T, Sugasaki T, Kawai S, Mita K, Oshiki T (2000) Molecular structure of a novel gypsy- Ty3-like retrotransposon (Kabuki) and nested retrotransposable elements on the W chromosome of the silkwom Bombyx mori. Mol Gen Genet 263:916–924
Boeke JD, Eichinger D, Castrillon D, Fink GR (1988) The Sacharomyces cerevisiae genome contains functional and non functional copies of transposon Ty1. Mol Cell Biol 8:1432–1442
Cáceres M, Ranz JM, Barbadilla A, Long M, Ruiz A (1999) Generation of a widespread Drosophila inversion by a transposable element. Science 285:415–418
Cañizares J, Grau M, Paricio N, Moltó MD (2000) Tirant is a new member of the gypsy family of retrotransposons in Drosophila melanogaster. Genome 43:9–14
Capy P, Vitalis R, Langin T, Higuet D, Bazin C (1996) Relationships between transposable elements based upon the integrase-transposase domains: is there a common ancestor? J Mol Evol 42:359–368
Casals F, Caceres M, Manfrin MH, González J, Ruiz A (2005) Molecular characterization and chromosomal distribution of Galileo, Kepler and Newton, three Foldback transposable elements of the Drosophila buzzatii species complex. Genetics 169:2047–2059
Christy RJ, Brown AR, Gourlie BB, Huang RCC (1985) Nucleotide sequences of murine intracisternal A-particle gene LTRs have extensive variability within the R region. Nucleic Acids Res 13:289–302
Conte C, Calco V, Desset S, Leblanc P, Dastugue B, Vaury C (2000) Impact of multiple insertions of two retroelements, Zam and Idefix at an euchromatic locus. Genetica 109:53–59
Covey SN (1986) Amino acid homology in gag regions of reverse transcribing elements and the coat protein gene of caulifower mosaic virus. Nucleic Acids Res 14:623–633
Doolittle RF, Feng DF, Johnson MS, McClure MA (1989) Origin and evolutionary relationships of retroviruses. Q Rev Biol 64:1–30
Evgen’ev MB, Corces VG, Lankenau DH (1992) Ulysses transposable element of Drosophila shows high structural similarities to functional domains of retroviruses. J Mol Biol 225:917–924
Evgen’ev MB, Zelentsova H, Poluectova H, Lyozin GT, Veleikodvorskaja V, Pyatkov KI, Zhivotovsky LA, Kidwell MG (2000) Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc Natl Acad Sci USA 10:11337–11342
Francino O, Cabré O, Fontdevila A (1993) Distribution of the copia transposable element in the repleta group of Drosophila. Genet Sel Evol 25:501–516
Friesen PD, Niessen M (1990) Gene organization and trascription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol 10:3067–3077
García Guerreiro MP, Fontdevila A (2001) Chromosomal distribution of the transposable elements Osvaldo and blanco in original and colonizer populations of Drosophila buzzatii. Genet Res 77:227–238
Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO (1999) Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92–104
Higgins DG, Thompson JD, Saigo K (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266:383–402
Hirsch MS, Curran J (1990) Human immunodeficiency viruses. In: Fields BN, Knipe DM (eds) Virology. Raven Press, New York
IHGSC (International Human Genome Sequencing Consortium) (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Inouye S, Yuki S, Saigo K (1986) Complete nucleotide sequence and genome organization of Drosophila transposable genetic element, 297. Eur J Biochem 154:417–425
Kaminker JSC, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, Ashburner M, Celniker SE (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3: Research0084
Ke N, Voytas DF (1997) High frequency cDNA recombination of the Saccharomyces retrotransposon Ty5: the LTR mediates formation of tandem elements. Genetics 147:545–556
Khan E, Mack JPG, Katz RK, Kulkosky J, Skalka AM (1990) Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res 4:851–860
Kolesnikov AV, Ponomarenko NA, Churikov NA (1996) Analysis of copies of the suffix-retrotransposon of Drosophila, produced using PCR on genomic DNA. Genetika 32:150–153
Kumar SK, Tamura IB, Jakobsen M, Nei M (2001) MEGA-2: Molecular Evolutionary Genetics Analysis software. Arizona, USA
Labrador M, Fontdevila A (1994) High transposition rates of Osvaldo, a new D. buzzatii retrotransposon. Mol Gen Genet 245:661–674
Labrador M, Naveira H, Fontdevila A (1990) Genetic mapping of the Adh locus in the repleta group of Drosophila by in situ hybridization. J Hered 81:83–86
Labrador M, Seleme MC, Fontdevila A (1998) The evolutionary history of D. buzzatii. XXXIV. The distribution of the retrotransposon Osvaldo in original an colonizer populations. Mol Biol Evol 15:1532–1547
Lankenau DH, Hauijser P, Jansen E, Miedema K, Henning W (1988) Micropia: a retrotransposon of Drosophila combining structural features of DNA viruses, retroviruses and non-viral transposable elements. J Mol Biol 204:233–246
Leblanc P, Desset S, Dastugue B, Vaury C (1997) Invertebrate retroviruses: ZAM a new candidate in D. melanogaster. EMBO J 16:7521–7531
Losada A, Abad JP, Villasante A (1997) Organization of DNA sequences near the centromere of the Drosophila melanogaster Y chromosome. Chromosoma 106:503–512
Maniatis TE, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Marin I, Labrador M, Fontdevila A (1992) The evolutionary history of Drosophila buzzatii. XXII. High content of nonsatellite repetitive DNA in D. buzzatii and its sibling D. koepferae. Genome 35:967–974
Marlor RL, Parkhust SM, Corces VG (1986) The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol 6:1129–1134
McClure MA (1991) Evolution of retrotransposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol 8:835–856
McClure MA (1992) Sequence analysis of eukaryotic retroid proteins. Math Comput Model 16:121–136
Mount SM, Rubin GM (1985) Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol 5:1630–1638
Mugnier N, Biémont C, Vieira C (2005) New regulatory regions of Drosophila 412 retrotransposable element generated by recombination. Mol Biol Evol 22:747–757
Nadir E, Margalit H, Gallil T, Ben-Sasson SA (1996) Microsatellite spreading in the human genome. Evolutionary mechanisms and structural implications. Proc Natl Acad Sci USA 93:6470–6475
Pantazidis A, Labrador M, Fontdevila A (1999) The retrotransposon Osvaldo from D. buzzatii displays all structural features of functional retrovirus. Mol Biol Evol 16:909–921
Piñol J, Francino O, Fontdevila A, Cabré O (1988) Rapid isolation of Drosophila high molecular weight DNA to obtain genomic libraries. Nucleic Acids Res 16:2736–2737
Priimaegi AF, Mizhrokhi LJ, Ilyin YV (1988) The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene 70:253–262
Ramsay L, Macaulay M, Cardle L, Morgante M, Ivanissevich S, Maestri E, Powell Waugh R (1999) Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J 17:415–425
Rein A, Henderson LE, Levin JG (1998) Nucleic-acid-cahaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci 23:297–301
Saigo K, Kugimiya W, Matsuo Y, Inouye S, Yoshioka K, Yuki S (1984) Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature 312:659–661
Saitou N, Nei M (1987) The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Schmidt ER (1992) A simplified and efficient protocol for non-radiactive in situ hybridisation to polytene chromosomes with a DIG.-labeled DNA probe. In: Non-radioactive in situ hybridisation application manual. Roche, pp 36–38
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
SanMiguel P, Gaut BS, Thikonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20:43–45
Tanda S, Mullor JL, Corces VG (1994) The Drosophila tom retrotransposon encodes an envelope protein. Mol Cell Biol 14:5392–5401
Temin HM (1981) Structure variation and synthesis of retrovirus long terminal repeats. Cell 27:1–3
Vincent JP, Girdham CH, O´Farrell PH (1994) A cell-autonomous ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev Biol 164:328–331
Vogt VM (1997) Retroviral virions and genomes. In: Coffin J, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory press, New York
Wasserman M (1982) Evolution of the repleta group. In: Ashburner M, Carlson HL, Thompson JN (eds) The genetics and biology of Drosophila. Academic, London, pp 62–139
Whalen JH, Grigliatti TA (1998) Molecular characterization of a retrotransposon in Drosophila melanogaster, nomad, and its relationship to other retrovirus-like mobile elements. Mol Gen Genet 260:401–409
Xiong Y, Eickbush TH (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362
Yuki S, Inouye S, Ishimaru S, Saigo K (1986) Nucleotide sequence characterization of a Drosophila retrotransposon, 412. Eur J Biochem 158:403–410
Zelentsova H, Poluectova H, Mnjoian L, Lyozin G, Veleikodvorskaja V, Zhivotovsky L, Kidwell MG, Evgen’ev MB (1999) Distribution and evolution of mobile elements in the virilis species group of Drosophila. Chromosoma 108:443–456
Acknowledgments
This research has been supported by grants BOS2000-0295-C02 and BOS2003-05904-C02 from Ministerio de Ciencia y Tecnología (MCYT) Spain and 2001SGR-00207 from the Generalitat de Catalunya to AF. We acknowledge C. Biémont and two anonymous reviewers for their helpful discussions and comments on the manuscript, and we thank L. Alarcón for providing us with DNA samples from different species of the buzzatii complex.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Reuter.
Rights and permissions
About this article
Cite this article
Guerreiro, M.P.G., Fontdevila, A. Molecular characterization and genomic distribution of Isis: a new retrotransposon of Drosophila buzzatii . Mol Genet Genomics 277, 83–95 (2007). https://doi.org/10.1007/s00438-006-0174-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0174-0