Abstract
Cystic echinococcosis is a worldwide zoonosis caused by Echinococcus granulosus. Because the methods of diagnosis and treatment for cystic echinococcosis were limited, it is still necessary to screen target proteins for the development of new anti-hydatidosis vaccine. In this study, the triosephosphate isomerase gene of E. granulosus was in silico cloned. The B cell and T cell epitopes were predicted by bioinformatics methods. The cDNA sequence of EgTIM was composition of 1094 base pairs, with an open reading frame of 753 base pairs. The deduced amino acid sequences were composed of 250 amino acids. Five cross-reactive epitopes, locating on 21aa-35aa, 43aa-57aa, 94aa-107aa, 115-129aa, and 164aa-183aa, could be expected to serve as candidate epitopes in the development of vaccine against E. granulosus. These results could provide bases for gene cloning, recombinant expression, and the designation of anti-hydatidosis vaccine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cystic echinococcosis (CE) is a worldwide zoonosis caused by Echinococcus granulosus. E. granulosus affects over one million people and is responsible for over $3 billion in expenses every year (Aqudelo Hiquita et al. 2016). The life cycle of E. granulosus is complex and involves two hosts, including an intermediate host and a definitive host. One of the strategies to reduce the risk of infection is to develop safe, effective, and economic anti-hydatidosis vaccine, which can interrupt the transmission of CE and reduce the risk to humans. Up to date, the effects of anti-hydatidosis vaccine are far from satisfactory. Therefore, it is still necessary to find immune markers which can be used in diagnosis and vaccine development.
Although E. granulosus uses both aerobic and anaerobic carbohydrate metabolisms to generate ATP, E. granulosus mainly depends on glycolysis for its energy supply (Cui et al. 2013). Triosephosphate isomerase (TIM) is a key regulatory enzyme of glycolysis, and was expressed in larval and adult development stages of E. granulosus and identified as antigenic protein by proteomic methods (Cui et al. 2013; Wang et al. 2015). TIM from Schistosoma japonicum (SjTIM) and Plasmodium falciparum (PfTIM) was identified as a target of drug or vaccine (Dai et al. 2014; Uiiah et al. 2012). TIM from other parasites have been cloned, recombinantly expressed, and analyzed by bioinformatics methods, including Fasciola hepatica, Clonorchis sinensis, and S. japonicum (Zinsser et al., 2013; Zhou et al., 2015; Zhang et al., 2015).
In silico cloning and prediction of B/T cell epitopes by bioinformatics is a new approach to search candidate vaccine molecule. Given that homologous genes from various species are conserved, in silico cloning can obtain partial or full-length sequences of complementary DNA (cDNA) by bioinformatics method, regardless of isolation among species. The principle of in silico cloning was that a sequenced gene from other species was used as a probe to search the expressed sequence tag (EST) database of target species. With the development of in silico cloning, we can find new methods to clone new genes and provide ideas for studies of gene function and proteomics. Currently, the studies on TIM of E. granulosus (EgTIM) are still scanty. In this study, EgTIM was in silico cloned. The sequence and coding protein characteristics were analyzed by bioinformatics methods. B cell and T cell epitopes of EgTIM were predicted by bioinformatics methods, providing references for the designation of anti-hydatidosis vaccine.
Materials and methods
In silico cloning of EgTIM
The protein sequence of SjTIM (accession number: AAC47393) was used as a probe to search the EST database of E. granulosus through the tblastn program in BLAST online software (http://blast.ncbi.nlm.nih.gov/) for a homologous search. The EST sequences of score more than 100 and query cover more than 50 % were used to generate contig through SeqMan program in DNAStar software. The above step was not stopped until the newly generated contig could not be elongated. These steps lead to a sequence as a putative cDNA sequence of EgTIM gene.
Bioinformatics analysis and B/T cell epitopes prediction of EgTIM
The open reading frame (ORF) and deduced amino acids sequences were analyzed by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/). The physiological biochemical characters of EgTIM, such as molecular weight, isoelectric point, and instability index were predicted by the ProtParam online software (http://web.Expasy.org/protparam/). Signal peptides were predicted by the SignalP 4.1 Server online software (http://www.cbs.dtu.dk/services/SignalP/). Post-translational modification sites were identified by the MotifScan online software (http://hits.isb-sib.ch/cgi-bin/motif_scan/). The structural domain was predicted by SMART (http://smart.embl-heidelberg.de/). The subcellular localization was predicted by ProtComp v. 9.0 (http://linux1.softberry.com/berry.phtml?topic=protcompan&group=programs&subgroup=proloc/). The transmembrane regions were predicted by TMPred (http://embnet.vital-it.ch/cgi-bin/TMPRED_form_parser). The hydrophilicity plot was predicted by the ProtScale (http://web.expasy.org/protscale/). DNAStar software was used to analyze the flexible regions, surface accessibility regions, and antigenic index. The secondary structures were predicted by SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma/). The T cell epitopes were analyzed by SYFPEITHI (http://www.syfpeithi.de/bin/MHCServer.dll/EpitopePrediction/). The 3-dimensional (3D) structures of EgTIM were constructed by the automated modeling program within the online service SWISS-MODEL. The 3D models of EgTIM were assessed by Verify_3D (http://services.mbi.ucla.edu/Verify_3D/). Two sequences alignment was analyzed by blast2p in NCBI (http:// http://blast.ncbi.nlm.nih.gov/BlastAlign.cgi/). Multiple-sequence alignment was analyzed by Clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was constructed by MEGA version 6.06. (http://www.megasoftware.net/).
Results
In silico cloning of EgTIM
With the protein sequence of SjTIM (accession number: AAC47393) as probe, 12 EST sequences of score more than 100 and query cover more than 50 % were found in the EST database of E. granulosus through the tblastn program in BLAST online software. These EST sequences were downloaded as FASTA format separately and assembled by SeqMan program (Fig. 1). The above step was repeated until the newly generated contig could not be elongated. One contig with the length of 1094 base pairs was obtained by SeqMan.
ORF and deduced amino acid sequences of EgTIM
The cDNA sequence was composed of 1094 base pairs. The ORF was located between 41 and 793 positions, with the length of 753 base pairs. The start codon was ATG and the stop codon was TGA. The deduced amino acid sequences were composed of 250 amino acids.
Physiological and biochemical characters of EgTIM
The number of amino acids was 250. The formula of EgTIM was C1205H1904N332O360S10, with a molecular weight of 27.12 kDa. The predicted isoelectric point of EgTIM was 6.60. EgTIM was predicted as a stable protein with the instability index of 22.14. The grand average of hydropathicity was −0.163, indicating that EgTIM was a hydrophilic protein.
Signal peptides and transmembrane regions of EgTIM
The result of signal peptides prediction showed that EgTIM had no signal peptides. The transmembrane regions were not found in EgTIM by TMPred either.
Subcellular localization of EgTIM
The subcellular localization of EgTIM was cytoplasmic.
Post-translational modification sites of EgTIM
One N-glycosylation site, three casein kinase II phosphorylation sites, five N-myristoylation sites, five protein kinase C phosphorylation sites, and one tyrosine kinase phosphorylation site were predicted in EgTIM (Table 1). Triosephosphate isomerase active site was located between 165 and 175 positions.
Structural domain of EgTIM
TIM family signature was found in EgTIM, located between 5 and 246 positions (Fig. 2).
Hydrophilicity/hydrophobicity of EgTIM
Multiple hydrophilic regions were found in EgTIM (Fig. 3).
The prediction of hydrophilicity/hydrophobicity of EgTIM. Vertical axis indicated hydrophobicity/hydrophilicity, and the horizontal axis indicated the position of amino acids in this protein. The positive value of the vertical axis indicated hydrophobicity and the negative value indicated hydrophilicity
Secondary structures of EgTIM
The secondary structures of EgTIM were consisted of α-helix, β-strands, β-turn, and random coil. The constituent ratio of these four secondary structures was 36 %, 22 %, 12 %, and 30 %, respectively.
Potential B cell epitopes of EgTIM
The flexible regions, surface accessibility regions, and antigenic index were predicted by Karplus-Schulz method, Emini method, and Jameson-Wolf index method, respectively. Six regions with good flexibility, surface accessibility, and high antigenic index were predicted in EgTIM, located on 25aa-35aa, 50aa-58aa, 94aa-98aa, 129aa-140aa, 152aa-158aa, and 172aa-183aa (Fig 4).
Flexibility (blue), surface accessibility (yellow), and antigenic index (red) of EgTIM. Horizontal axis indicated the position of amino acids in this protein, and the vertical axis indicated flexibility index, surface accessibility index, or antigenic index. The zones above the horizontal axis indicated the potential B cell epitopes
T cell epitopes of EgTIM
Cytotoxic T cell (CTL) epitopes with the length of 9aa were analyzed by the selection of HLA-A*0201 within the online service SYFPEITHI. Eight CTL epitopes with the score of more than 21 were predicted in EgTIM (Table 2). Helper T cell (Th) epitopes with the length of 15aa were predicted by the selection of HLA-DRB1*0401 within SYFPEITHI. Eleven Th cell epitopes with the score of more than 20 were predicted in EgTIM (Table 3).
Homology modeling of EgTIM
The template sequence of triosephosphate isomerase of Rhipicephalus microplus (RmTIM) (PDB code: 3th6.1.A) was used to construct 3D structures of EgTIM homodimeric molecule, with a sequence identity of 67.14 %. Characteristic “barrel”-shaped formed from β-strands and conserved triosephosphate isomerase active sites were found in EgTIM (Fig. 5). Analysis of the quality of EgTIM using Verify 3D online software showed that 97.69 % of the residues had an averaged 3D/1D sore ≥0.2 (more than 65 %), indicating that homology modeling of EgTIM was reliable.
Multiple-sequence alignment and the construction of phylogenetic tree
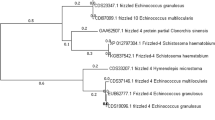
The results of multiple-sequence alignments showed that the triosephosphate isomerase active site sequences (AYEPVWAIGTG) were conserved among all aligned species with the exception of Giardia lamblia (G. lamblia) and P. falciparum (Fig. 6). The results of two-sequence alignments showed that EgTIM share the highest sequence identity with TIM of Taenia solium (TsTIM, 94 %). The sequence identity between EgTIM and PfTIM was the lowest (41 %). EgTIM clustered with TsTIM, which belonged to tapeworm. TIM from human and Mus musculus (M. musculus) were well separated from EgTIM (Fig. 7).
Multiple-sequence alignments of TIM from T. solium (accession number AAG21132); Trypanosoma cruzi (accession number AAB58349); Giardia lamblia (accession number P36186); Clonorchis sinensis (accession number GAA50994); Schistosoma japonicum (accession number AAC47393); Schistosoma mansoni (accession number AAA29919); Plasmodium falciparum (accession number AAA18799); Toxoplasma gondii (accession number ABE76515); Fasciola hepatica (accession number: AGJ83762); Brugia malayi (accession number XP_001897269); Mus musculus (accession number CAA37420); Homo sapiens (accession number CAA49379); and E. granulosus. The frames indicated triosephosphate isomerase active sites
Phylogenetic tree constructed using the neighbor-joining method to compare the relationship between EgTIM and TIM from other species. The numbers above the branches refer to bootstrap values. The accession numbers and species for sequences included in the phylogenetic analysis are shown behind the branches
Discussion
Cystic echinococcosis (CE), a worldwide zoonosis caused by E. granulosus, affects over one million people every year (Aqudelo Hiquita et al. 2016). The diagnosis and treatment were still limited, therefore, it is necessary to screen target proteins for the development of new anti-hydatidosis vaccine. Screening the genes involved in energy metabolism or immune response from hydatid genome database and evaluating their values in vaccine researches have become a new research focus. With the development of genomics and proteomics, conventional experimental methods can not meet the needs of screening of candidate vaccine. In silico cloning and prediction of B/T cell epitopes by bioinformatics accelerated the screening of candidate vaccine molecule, providing bases for gene cloning, recombinant expression, and the development of peptide vaccine. In silico cloning has been used to clone functional genes from various species, including human, non-amniote vertebrates, Carica papaya (C. papaya), Chlamydomonas reinhardtii (C. reinhardtii), dsRNA viruses, spider mite and Trypanosoma brucei. These genes have been identified by reverse transcription PCR (RT-PCR) or further researches, indicating the validity of in silico cloning (Obrien et al. 2000; Zhang et al. 2013; Idrove Espin et al. 2012; Herrera-Valencia et al. 2012; Liu et al. 2012; Veenstra et al. 2012; Akhoon et al. 2011). The highly conserved and specific epitopes of VSG from T. brucei have been identified by in silico cloning and, these epitopes were used to design a potential vaccine, which was found to be effective even for non-African populations (Akhoon et al. 2011). These results indicated that in silico cloning may serve as a new method to develop new vaccine.
More than 5000 EST sequences were collected in EST database of E. granulosus, providing bases for in silico cloning. The sequence of SjTIM had been cloned and sequenced (Zhang et al. 2015). In this study, a full-length cDNA sequence of EgTIM was obtained with the probe of SjTIM since both parasites are helminthes, and most platyhelminth may share some similar antigens. The cDNA sequence of EgTIM was composed of 1094 base pairs, with an ORF of 753 base pairs. The deduced amino acid sequences were composed of 250 amino acids. Triosephosphate isomerase active site sequences (AYEPVWAIGTG) were conserved among all aligned species with the exception of G. lamblia and P. falciparum. EgTIM clustered with TsTIM, which belonged to tapeworm of the same family Taeniidae. TIM from human and M. musculus were well separated from EgTIM. The results of phylogenetic tree construction were consistent with traditional categories. These results indicated that the validity of the obtained cDNA sequence was high.
Epitopes can bind to antibody specifically and located to antigen molecule surface, including B cell epitopes and T cell epitopes. In the prediction of B cell epitopes, the results of hydrophilicity, flexibility, surface accessibility, antigenic index, and secondary structures were considered. The hydrophilicity, flexibility, surface accessibility, and antigenic index of epitopes were high (Cui et al. 2015). Multiple hydrophilic regions were found in EgTIM. α-Helix and random coil were the main secondary structures of EgTIM, indicating that multiple regions of EgTIM can serve as potential epitopes. In this study, six regions with good flexibility, surface accessibility, and high antigenic index were predicted in EgTIM, located on 25aa-35aa, 50aa-58aa, 94aa-98aa, 129aa-140aa, 152aa-158aa, and 172aa-183aa.
T cell epitopes were divided into CTL epitopes and Th cell epitopes, referring to linear peptides presented to T cell receptor by MHC molecules. CTL epitopes were restricted by MHC I molecules, and Th cell epitopes were restricted by MHC II molecules. In this study, eight CTL epitopes and 11 Th cell epitopes were found in EgTIM. It is noteworthy that triosephosphate isomerase active site was included partially or fully in CTL epitopes, located on 164aa-172aa, and Th cell epitopes, located on 166aa-180aa, and 163aa-177aa.
Cross-reactive epitopes were referring to the peptides combined with T and B cell epitopes, which can induce not only humoral immune but also cellular immune (Flower 2013). Integrating consideration of the results of T cell and B cell epitope prediction, a total of five cross-reactive epitopes were found in EgTIM, located on 21aa-35aa, 43aa-57aa, 94aa-107aa, 115-129aa, and 164aa-183aa. Taken together, EgTIM may serve as a candidate antigen for the development of vaccine, because of good antigenicity.
The prediction of 3D structures of protein can provide theoretical references for the designation of epitopes vaccine, the study of antibody variable domains, and the screening of target proteins. The homology modeling method has proved to be one of the most robust modeling tools. Given that the sequence identity between template and target protein was more than 60 %, the predicted 3D structure was accurate and close to the experimental results. In this study, RmTIM was used as a template to construct 3D structure of EgTIM homodimeric, which shared a sequence identity of 67.14 % with EgTIM. Analysis of the quality of EgTIM using Verify 3D online software showed that 97.69 % of the residues had an averaged 3D/1D sore ≥0.2 (more than 65 %), indicating homology modeling of EgTIM was reliable.
In conclusion, a full-length cDNA sequence of EgTIM was obtained by the method of in silico cloning. The cDNA sequence was composed of 1094 base pairs, with a ORF of 753 base pairs. Five cross-reactive epitopes, located on 21aa-35aa, 43aa-57aa, 94aa-107aa, 115-129aa, and 164aa-183aa, could be expected to serve as candidate epitopes in the development of vaccine against E. granulosus. The accuracy of in silico cloning was restricted by the quality of EST database. The cDNA sequences obtained from in silico cloning must be identified by further experimental methods.
References
Akhoon BA, Slathia PS, Sharma P, Gupta SK, Verma V (2011) In silico identification of novel protective VSG antigens expressed by Trypanosoma brucei and an effort for designing a highly immunogenic DNA vaccine using IL-2 as adjuvant. Microb Pathog 51:77–87
Aqudelo Hiquita NI, Brunetti E, McCloskey C (2016) Cystic echinococcosis. J Clin Microbiol 3:518–523
Cui SJ, Xu LL, Zhang T, Xu M, Yao J, Fang CY, Zheng F, Yang PY, Wei H, Liu F (2013) Proteomic characterization of larval and adult developmental stages in Echinococcus granulosus reveals novel insight into host-parasite interactions. J Proteome 84:158–175
Cui JM, Zhang NZ, Li WH, Yan HB, Fu BQ (2015) Cloning, identification, and bioinformatics analysis of a putative aquaporin TsAQP from Trichinella spiralis. Genet Mol Res 14:12699–12709
Dai Y, Wang XT, Zhao S, Tang JX, Zhang L, Dai JR, Zeng MT, Lu S, Zhu YC, Su C (2014) Construction and evaluation of replication-defective recombinant optimized triosephosphate isomerase adenoviral vaccination in Schistosoma japonicum challenged mice. Vaccine 32:771–778
Flower DR (2013) Designing immunogenic peptides. Nat Chem Biol 9:749–753
Herrera-Valencia VA, Macario-Gonzalez LA, Molina-Casais ML, Beltran-Aguilar AG, Peraza-Echeverria S (2012) In silico cloning and characterization of the glycerol-3-phosphate dehydrogenase (GPDH) gene family in the green microalga Chlamydomonas reinhardtii. Curr Microbiol 64:477–485
Idrove Espin FM, Peraza-Echeverria S, Fuentes G, Santamaria JM (2012) In silico cloning and characterization of TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiol Biochem 54:113–122
Liu HQ, Fu YP, Xie JT, Cheng JS, Ghabrial SA, Li GQ, Yi XH, Jiang DH (2012) Discovery of novel dsRNA viral sequences by in silico cloning and implications for viral diversity, host range and evolution. PLoS ONE 7:e42147
Obrien KP, Tapia-Paez I, Stahle-Backdahl M, Kedra D, Dumanski JP (2000) Characterization of five novel human genes in 11q13-q22 region. Biochem Biophys Res Commun 273:90–94
Uiiah M, Hira J, Ghosh T, Ishaque N, Absar N (2012) A bioinformatics approach for homology modeling and binding site identification of triosephosphate isomerase from Plasmodium falciparum 3D7. J Young Pharm 4:261–266
Veenstra JA, Rombauts S, Grbic M (2012) In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem Mol Biol 42:277–295
Wang Y, Xiao D, Shen YJ, Han XM, Zhao F, Li XH, Wu WP, Zhou HJ, Zhang JZ, Cao JP (2015) Proteomic analysis of the excretory/secretory products and antigenic proteins of Echinococcus granulosus adult worms from infected dogs. BMC Vet Res 11:119–125
Zhang YB, Liu TK, Jiang J, Shi J, Liu Y, Li S, Gui JF (2013) Identification of a novel Gig2 gene family specific to non-amniote vertebrates. PLoS ONE 8:e60588
Zhang W, Luo X, Zhang F, Zhu Y, Yang B, Hou M, Xu Z, Yu C, Chen Y, Chen L, Ji M (2015) SjTat-TPI facilitates adaptive T-cell responses and reduces hepatic pathology furing Schistosoma japonicum infection in BALB/c mice. Parasit Vectors 8:664–672
Zhou JJ, Liao H, Li S, Zhou CH, Huang Y, Li XR, Liang C, Yu XB (2015) Molecular identification, immunolocalization, and characterization of Clonorchis sinensis triosephosphate isomerase. Parasitol Res 114:3117–3124
Zinsser VL, Hoey EM, Trudgett A, Timson DJ (2013) Biochemical characterization of triose phosphate isomerase from the liver fluke Fasciola hepatica. Biochimie 95:2182–2189
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers 30972567 and 30371258).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, F., Ye, B. In silico cloning and B/T cell epitope prediction of triosephosphate isomerase from Echinococcus granulosus . Parasitol Res 115, 3991–3998 (2016). https://doi.org/10.1007/s00436-016-5166-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-016-5166-3