Abstract
The Plasmodium vivax merozoite surface protein 1 (Pvmsp-1) locus codes for a major asexual blood-stage antigen currently proposed as a malaria vaccine candidate antigen. However, extensive polymorphism of this protein has been observed in isolates from different geographical areas. Here, we investigate the extent and the frequency of allelic diversity at the Pvmsp-1 locus in field isolates collected in the Republic of Korea during the past decade. Among the 45 Korean isolates, six Pvmsp-1 gene types (SKOR-I to SKOR-VI) were identified as unique combinations of type sequences in each variable block. Of these six different Pvmsp-1 gene types, two major Pvmsp-1 allelic types were found in 72% (SKOR-I) and 28% (SKOR-II) of field isolates collected in 1996 to 2000, and four different allelic types (SKOR-III to SKOR-VI) emerged in 70% (10–25%) of isolates collected in 2007 to 2009. These results suggest that allelic diversity of Pvmsp-1 increased in several variable regions, including the N- and C-terminals, after reemergence of P. vivax parasites in the Republic of Korea.
Similar content being viewed by others
Introduction
Plasmodium vivax is the most widely distributed species that causes human malaria. With 80 to 300 million clinical cases reported each year, P. vivax presents a major public health challenge in Central and South America, the Middle East, Central, South, and Southeast Asia, Oceania, and East Africa (Mendis et al. 2001; Mueller et al. 2009). The complex biology of the P. vivax parasites and its transmitting vectors has hindered the development of successful control strategies. The long-term goal of P. vivax prevention will require a combined approach that includes vaccination, chemotherapy, and vector control (Mendis et al. 2001).
Understanding of the extent and the dynamics of genetic diversity in target antigens is needed for the development of new vaccines and the interpretation of the result of the vaccine efficacy tests. Because amino acid variations caused by single-point mutations can affect the immunogenic properties of antigens, identification of genetic polymorphisms of target antigenic domains is essential for vaccine development (Takala and Plowe 2009). Several laboratory and epidemiological studies have demonstrated that natural variations can abrogate immune recognition in Plasmodium species (Tonhosolo et al. 2001; Tonon et al. 2004; Bastos et al. 2007; Mehrizi et al. 2009). Thus, the investigation of sequence polymorphism and antigenic diversity of malaria vaccine candidate antigens has become a subject of considerable importance.
P. vivax merozoite surface protein 1 (PvMSP-1) is a 200-kDa polymorphic glycoprotein (Udagama et al. 1990) whose complete gene sequence is firstly characterized in two Latin American isolates, the Brazilian Belem (Belem) and Salvador (Sal-1) strains (Del Portillo et al. 1991; Gibson et al. 1992). The PvMSP-1 has been shown to produce naturally acquired and vaccine-induced immunity and has thus been identified as an important vaccine candidate among PvMSPs (PvMSP-1, -4, -5, -8, and -10) (Barrero et al. 2005; Dutta et al. 2005). Several experimental studies using native and recombinant PvMSP-1, particularly its C-terminal 19-kDa fragment, have demonstrated the vaccine potential of this protein (Galinski and Barnwell 2008). The Pvmsp-1 gene has a mosaic and heterogeneity structure with allelic recombination demonstrated in isolates from different countries (Putaporntip et al. 2002). This gene also contains several conserved blocks (CBs) and variable blocks (VBs) (Fig. 1a; del Portillo et al. 1991; Putaporntip et al. 2002). The VB6 region between CB5 and CB6 shows a dimorphic pattern of “Belem type” and “Sal-1 type” strains. Interallelic recombination between these types occurs at a high frequency among worldwide isolates (Kolakovich et al. 1996; Putaporntip et al. 1997; Figtree et al. 2000; Lim et al. 2000; Severini et al. 2002; Cui et al. 2003; Zakeri et al. 2006). Because Pvmsp-1 not only encodes a strong vaccine candidate, but is also a useful marker for the genetic polymorphism of the parasite, we considered it important to determine the natural variation and prevalence of Pvmsp-1 alleles.
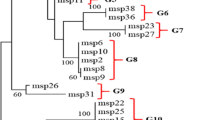
Schematic representation of the merozoite surface protein-1 gene of Plasmodium vivax (a) and its association with the type sequence of each variable block in this study (b). a Conserved and variable blocks of the gene are shown as open and closed boxes, respectively. b Schematic representation of six allelic types (SKOR-I to SKOR-VI) and their associations of type sequences in each variable block from the Pvmsp-1 gene. Variable blocks 2, 6, 8, and 10 contain short tandem repeats, indicated by number. Detailed allelic type and amino acid sequence alignment data are shown in Table 2 and Supplementary Fig. 1. Box numbers and the sequence type of each variable block are based on a previous report (Putaporntip et al. 2002)
In the present study, we determined the frequencies and the genetic variation of Pvmsp-1 alleles by analyzing full-length sequences from field isolates of reemergent P. vivax collected in the Republic of Korea.
Materials and methods
Blood samples and DNA preparation
Blood samples were collected from 45 symptomatic patients (25 from 1996 to 2000, 20 from 2006 to 2009) who had been diagnosed with P. vivax infection by microscopic examination at local health centers and clinics in Gyeonggi and Gangwon Provinces, Republic of Korea. This study was approved by the Institutional Review Board of Kangwon National University Hospital. Genomic DNA was purified from 200 μL of the whole blood using QIAamp DNA Blood Mini Kits (Qiagen, Seoul, Korea), according to the manufacturer’s instructions. The purified DNA was dissolved in TE buffer (10 mM Tris–HCl, 1 mM EDTA; pH 8.0) and stored at −20°C until use.
Polymerase chain reaction and sequencing
The DNA fragment spanning the entire coding region of the Pvmsp-1 gene was amplified using primers based on the Pvmsp-1 sequence of the Sal-1 strain (Putaporntip et al. 2002). Polymerase chain reaction (PCR) amplification was performed by 35 cycles of denaturing the samples at 94°C for 30 s, annealing at 60°C for 30 s, and extending at 72°C for 5 min. To minimize errors introduced in the sequences during PCR amplification, we used ExTaq and LATaq DNA polymerase (Takara, Shiga, Japan). The PCR products were examined by electrophoresis in a 1% agarose gel, visualized with an ultraviolet transilluminator, purified with PCR purification kits (Qiagen), and both strands sequenced using a BigDye Terminator Cycle Sequencing Kit on an ABI3100 genetic analyzer (Applied Biosystems, Foster City, CA) using previously reported forward and reverse primers (Putaporntip et al. 2002). When a singleton occurred, the sequence was redetermined using DNA templates from two independent amplifications from the same DNA samples.
Sequence analysis
Preliminary pairwise sequence alignments and comparisons were performed using MegAlign (DNASTAR, Madison, WI). A multiple-sequence alignment was constructed with the MUSCLE (Edgar 2004) and BioEdit programs (Ibis Therapeutics, Carlsbad, CA). The Pvmsp-1 gene sequences of the Korean isolates in this study were then compared with previously reported sequences retrieved from GenBank using published accession numbers (Putaporntip et al. 2002). Sequences obtained in this study have been deposited in GenBank with the accession numbers HQ171934–HQ171941.
Results
The frequency of Pvmsp-1 allelic types and the association of type sequences in each VB were studied and are shown in Table 1 and Fig. 1. To assess the genetic diversity of Korean P. vivax isolates at the Pvmsp-1 locus, we analyzed the Pvmsp-1 sequences of 45 clinical samples from P. vivax-endemic areas in the Republic of Korea. Comparison with previously reported (Putaporntip et al. 2002) Pvmsp-1 sequences from the Sal-1 and Belem strains allowed us to identify six allelic types among the Korean isolates (Fig. 1). Two major allelic types (SKOR-I and SKOR-II) were prevalent among the Korean isolates collected from 1996 to 2000, accounting for 72.0% (SKOR-I) and 28.0% (SKOR-II) of the samples. Six allelic types (SKOR-I to SKOR-VI) occurred with similar frequency (average 16.7%, range 10–25%) in the Korean isolates collected from 2007 to 2009, and four new allelic types (SKOR-III to SKOR-VI) were identified. Together, these four types comprised approximately 70% (10–25% each) of the isolates (Table 1).
Two to five type sequences from each allelic type were identified in each VB of the Korean isolates. All the VB regions in SKOR-I and SKOR-II except two (VB2b and VB10) contained a single type sequence. In contrast, the VBs in SKOR-III to SKOR-VI contained two or three new type sequences (Table 2). In samples collected from 2007 to 2009, most of the type sequence in each VB differed from those in the samples collected from 1996 to 2000. SKOR-III and SKOR-IV contained the same type sequence in five VBs (2a, 2c, 4, 6, and 8) and different type sequences in three VBs (2b, 10, and 12; Fig. 1b). SKOR-I was similar to SKOR-II, SKOR-III, SKOR-IV, SKOR-V, and SKOR-VI, with 97.8%, 92.5%, 90.4%, 85.2%, and 85.5% sequence similarity, respectively.
VBs 2, 6, 8, and 10 contained short tandem repeat (STR) regions (Supplementary Fig. 1). The degenerate 5-mer (GSXXX) n (n = 2, 3, 6, and 7) was found in VB2b; this block consisted of (GSXXX)7 in samples collected from 1996 to 2000 (types 5 and 7), but (GSXXX)2, 3, 6 in samples collected from 2007 to 2009 (types 10, 18, and 20). In VB6, polyglutamine (Q n ) (n = 1, 2, 10, and 14) was found in types 1 (and its variant), 2, and 4 sequences. The amino acid sequence of type 1 and its variant in VB6 were similar to that of the Sal-1 type, with few Q repeats, and the sequence of type 2 was similar to that of Belem, with multiple Q repeats. The sequence of type 4 (recombinant type) in VB6 represents a combination of a Sal-1-like sequence at the 5′-end and a Belem-like sequence at the 3′-end, including the poly-Q segment. This sequence was found in all 25 sequences obtained from 1996 to 2000 and in six (30%) of the 20 sequences obtained from 2007 to 2009. The type 1 and type 2 sequences were found at the same frequency (7/20, 35%) among the samples collected from 2007 to 2009. In VB8, degenerate repeats of (PVTTTX)2 and (PXVAPX)2 plus PVTTNT were found in types 3 and 5, respectively. Type 5 was observed in 70% (13 isolates) of the allelic types SKOR-I to SKOR-IV collected from 2007 to 2009 and in 100% (25) of those collected from 1996 to 2000. The degenerate 5-mer (VPXXX) n (n = 2, 3, 5) was observed in VB10. Both type 10 and type 14 in VB10 were highly prevalent (75.6%), and other types appeared with two or three tandem repeat sequences. No distinctive repeat appeared in VB4 or VB12. In VB12, the C-terminal part of Pvmsp-1, three new variant types appeared after 2000.
The alignment of the Pvmsp-1 alleles from the Korean isolates revealed that amino acid sequences were highly conserved in the CBs (Supplementary Fig. 2). We summarized the nonsynonymous substitutions from six Pvmsp-1 CBs and observed the following unique sequences in the Korean isolates: SQ/TEA in CB1, NVSASSTQ/NVSASSTQ/EK/QA in CB3, SNTL in CB5, D in CB7, and AE/G in CB9. The sequence analysis of the C-terminal region (CB13) of the Pvmsp-1 gene showed no variation in Korean P. vivax and was identical to those of the Sal-1 and Belem strains. Many nonsynonymous mutation sites (Supplementary Fig. 2) in CB1, CB7, and CB9 were found in only two allelic types (SKOR0804 and SKOR0814) and were identical to the Belem type.
Discussion
In the present study, we investigated the allelic diversity of each block of full-length Pvmsp-1 from reemergent P. vivax parasites collected in the Republic of Korea during the past decade to understand the distribution of their genetic polymorphism. Conserved and variable blocks of the full-length Pvmsp-1 were compared among samples collected from periods both immediately following reemergence and about 10 years thereafter to determine whether the genetic diversity of P. vivax parasites had increased during this time. Our data identified four new Pvmsp-1 allelic types, indicating that the frequency of genetic diversity of this gene was increased in areas of malaria endemicity after the reemergence of P. vivax in Korea. It is very important to consider that longitudinal sampling as shown in this study is very rare in a low-endemicity country such as Korea with reemerging P. vivax and that it is valuable for understanding how the parasite population changes in the field.
Since the 1993 reemergence of P. vivax in the Republic of Korea, at least two genotypes have been found in vaccine candidate genes Pvmsp-1 (both ICB4–5 and ICB5–6), circumsporozoite protein (Pvcsp), Duffy binding protein (Pvdbp), and apical membrane protein 1 (Pvama-1), all of which were identified in Korean isolates obtained before 2000 (Lim et al. 2000; Han et al. 2002; Kim et al. 2009; Lim et al. 2001). In addition, subtypes or recombinant types have been found in recent sequence analyses of Pvmsp-1 ICB5–6, Pvmsp-3α, and Pvcsp in samples collected from 1996 to 2007 (Choi et al. 2010; Nam et al. 2010). In a previous report (Putaporntip et al. 2002), two Korean allelic types (SK1 and SK3) over the full length of Pvmsp-1 were identified from four Korean isolates (SK1 to SK4). These were identified from samples collected before 2000, and their VBs showed unique sequence types identical to some of the 31 types from isolates collected from worldwide. The present study identified four new allelic types in the Korean isolates collected from 2007 to 2009, with different type sequences associated with each VB and different numbers of STR sequences. These four allelic types were also identified from the unique association of each VB with STRs from isolates collected from worldwide. Although sequences from part of each allelic type were similar to those of isolates collected worldwide, each association of type sequences from the VBs differed from those previously reported.
Previous studies have examined the genetic diversity of Pvmsp-1 ICB5–6 in Columbia, Brazil, Myanmar, and Azerbaijan, all of which are high-risk regions for malaria (Leclerc et al. 2004; Maestre et al. 2004; Santos-Ciminera et al. 2007; Moon et al. 2009). These endemic geographical areas have emerged as an important source of genetic markers of P. vivax polymorphism that allows the detection of mixed infections and recombination events between the parental Sal-1 and Belem type alleles. In particular, high diversification has been observed in the variable poly-Q region between ICB5 and ICB6 of Pvmsp-1, where genetic recombination often occurs. Until recently, the number of type sequences in VB6 (similar to the ICB5–6 region) seemed to be restricted to four. These four type sequences (type 1 and variant, and types 2 and 4) of Pvmsp-1 VB6 from the Korean isolates from this study were similar to the previously reported S-a, S-b, B-1, and B-2, respectively (Choi et al. 2010). Although this VB sequence was found to be a major recombinant type before 2000, Sal-1- and Belem-like types appeared in the Korean isolates only after 2000 (Choi et al. 2010). In this study, a Sal-1-like type in VB2b of Pvmsp-1 was a major allelic type before 2000 and a Belem-like type of VB2a was found in the 2007 to 2009 samples. Sal-1 and Belem types of Pvmsp-1were predominant in VB2a and VB12 before 2000, but both basic and recombinant types were also observed in 2007 to 2009 samples. These findings indicate that a recombination independently changed the allelic type of each VB of Pvmsp-1 during the endemic phase of this parasite in the Republic of Korea.
Recombinant proteins corresponding to PvMSP-1 variable domains commonly found in field parasites were poorly recognized by normal populations who are exposed to malaria in endemic areas (Bastos et al. 2007). From this previous report, few subjects had detectable IgG antibodies against one or more of the several VBs 2, 6, and 10 expressed as recombinant proteins, although most of them recognized the conserved C-terminal domains of PvMSP-1. This CB10 contains MSP-1 19 kDa, which has been the focus of malaria vaccine development because of its highly conserved sequence and hypothesized critical function. The report indicated that sequence polymorphism affected the analysis of antibody recognition of PvMSP-1 variants, and this information can be used to understand naturally acquired immunity in the endemic population prior to the selection of a particular domain for vaccine development.
In conclusion, sequence analysis of the full-length Pvmsp-1 gene indicated that its genetic diversity has increased during the past decade from two genotypes to six. Monitoring of the genetic variation patterns of the P. vivax parasites may help to analyze its endemicity in the Republic of Korea. As allelic polymorphism is one of the greatest hurdles in mounting a protective immune response against the genetically diverse P. vivax, further research is required to determine the impact of antigenic diversification on the immunogenicity of the PvMSP-1 antigen in natural populations.
References
Barrero CA, Delgado G, Sierra AY, Silva Y, Parra-Lopez C, Patarroyo MA (2005) Gamma interferon levels and antibody production induced by two PvMSP-1 recombinant polypeptides are associated with protective immunity against P. vivax in Aotus monkeys. Vaccine 23:4048–4053
Bastos MS, da Silva-Nunes M, Malafronte RS, Hoffmann EH, Wunderlich G, Moraes SL, Ferreira MU (2007) Antigenic polymorphism and naturally acquired antibodies to Plasmodium vivax merozoite surface protein 1 in rural Amazonians. Clin Vaccine Immunol 14:1249–1259
Choi YK, Choi KM, Park MH, Lee EG, Kim YJ, Lee BC, Cho SH, Rhie HG, Lee HS, Yu JR, Lee JS, Kim TS, Kim JY (2010) Rapid dissemination of newly introduced Plasmodium vivax genotypes in South Korea. Am J Trop Med Hyg 82:426–432
Cui L, Escalante AA, Imwong M, Snounou G (2003) The genetic diversity of Plasmodium vivax populations. Trends Parasitol 19:220–226
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
del Portillo HA, Longacre S, Khouri E, David PH (1991) Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA 88:4030–4034
Dutta S, Kaushal DC, Ware LA, Puri SK, Kaushal NA, Narula A, Upadhyaya DS, Lanar DE (2005) Merozoite surface protein 1 of Plasmodium vivax induces a protective response against Plasmodium cynomolgi challenge in rhesus monkeys. Infect Immun 73:5936–5944
Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A (2000) Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA 1 and MSP 1 genes. Mol Biochem Parasitol 108:53–66
Galinski MR, Barnwell JW (2008) Plasmodium vivax: who cares? Malar J 7(1):S9
Gibson HL, Tucker JE, Kaslow DC, Krettli AU, CollinsWE KMC, Bathurst IC, Barr PJ (1992) Structure and expression of the gene for Pv200, a major blood-stage surface antigen of Plasmodium vivax. Mol Biochem Parasitol 50:325–334
Han ET, Park JH, Shin EH, Choi MH, Oh MD, Chai JY (2002) Apical membrane antigen-1 (AMA-1) gene sequences of re-emerging Plasmodium vivax in South Korea. Korean J Parasitol 40:157–162
Kim SH, Hwang SY, Shin JH, Moon CS, Kim DW, Kho WG (2009) Molecular genetic characterization of the merozoite surface protein 1 gene of Plasmodium vivax from reemerging Korean isolates. Clin Vaccine Immunol 16:733–738
Kolakovich KA, Ssengoba A, Wojcik K, Tsuboi T, Al-Yaman F, Alpers M, Adams JH (1996) Plasmodium vivax: favored gene frequencies of the merozoite surface protein-1 and the multiplicity of infection in a malaria endemic region. Exp Parasitol 83:11–19
Leclerc MC, Menegon M, Cligny A, Noyer JL, Mammadov S, Aliyev N, Gasimov E, Majori G, Severini C (2004) Genetic diversity of Plasmodium vivax isolates from Azerbaijan. Malar J 3:40
Lim CS, Kim SH, Kwon SI, Song JW, Song KJ, Lee KN (2000) Analysis of Plasmodium vivax merozoite surface protein-1 gene sequences from resurgent Korean isolates. Am J Trop Med Hyg 62:261–265
Lim CS, Kim YK, Lee KN, Kim SH, Hoffman KJ, Song KJ, Song JW (2001) The analysis of circumsporozoite-protein gene sequences from South Korean isolates of Plasmodium vivax. Ann Trop Med Parasitol 95:229–235
Maestre A, Sunil S, Ahmad G, Mohmmed A, Echeverri M, Corredor M, Blair S, Chauhan VS, Malhotra P (2004) Inter-allelic recombination in the Plasmodium vivax merozoite surface protein 1 gene among Indian and Colombian isolates. Malar J 3:4
Mehrizi AA, Zakeri S, Salmanian AH, Sanati MH, Djadid ND (2009) IgG subclasses pattern and high-avidity antibody to the C-terminal region of merozoite surface protein 1 of Plasmodium vivax in an unstable hypoendemic region in Iran. Acta Trop 112:1–7
Mendis K, Sina BJ, Marchesini P, Carter R (2001) The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg 64:97–106
Moon SU, Lee HW, Kim JY, Na BK, Cho SH, Lin K, Sohn WM, Kim TS (2009) High frequency of genetic diversity of Plasmodium vivax field isolates in Myanmar. Acta Trop 109:30–36
Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA (2009) Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis 9:555–566
Nam DH, Oh JS, Nam MH, Park HC, Lim CS, Lee WJ, Sattabongkot J, Klein TA, Ayala FJ (2010) Emergence of new alleles of the MSP-3alpha gene in Plasmodium vivax isolates from Korea. Am J Trop Med Hyg 82:522–524
Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho WG, Kaneko A, Kanbara H, Hattori T, Tanabe K (2002) Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc Natl Acad Sci USA 99:16348–16353
Putaporntip C, Jongwutiwes S, Tanabe K, Thaithong S (1997) Interallelic recombination in the merozoite surface protein 1 (MSP-1) gene of Plasmodium vivax from Thai isolates. Mol Biochem Parasitol 84:49–56
Santos-Ciminera PD, Alecrim MG, Roberts DR, Quinnan GV Jr (2007) Molecular epidemiology of Plasmodium vivax in the State of Amazonas, Brazil. Acta Trop 102:38–46
Severini C, Menegon M, Gradoni L, Majori G (2002) Use of the Plasmodium vivax merozoite surface protein 1 gene sequence analysis in the investigation of an introduced malaria case in Italy. Acta Trop 84:151–157
Takala SL, Plowe CV (2009) Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria’. Parasite Immunol 31:560–573
Tonhosolo R, Wunderlich G, Ferreira MU (2001) Differential antibody recognition of four allelic variants of the merozoite surface protein-2 (MSP-2) of Plasmodium falciparum. J Eukaryot Microbiol 48:556–564
Tonon AP, Hoffmann EH, Silveira LA, Ribeiro AG, Gonçalves CR, Ribolla PE, Wunderlich G, Ferreira MU (2004) Plasmodium falciparum: sequence diversity and antibody recognition of the merozoite surface protein-2 (MSP-2) in Brazilian Amazonia. Exp Parasitol 108:114–125
Udagama PV, Gamage-Mendis AC, David PH, Peiris JSM, Pepera KLRL, Mendis KN, Carter R (1990) Genetic complexity of Plasmodium vivax parasites in individual human infections analysed with monoclonal antibodies against variant epitopes on a single parasite protein. Am J Trop Med Hyg 42:104–110
Zakeri S, Mehrizi AA, Mamaghani S, Noorizadeh S, Snounou G, Djadid ND (2006) Population structure analysis of Plasmodium vivax in areas of Iran with different malaria endemicity. Am J Trop Med Hyg 74:394–400
Acknowledgments
This work was supported by a Korean Science and Engineering Foundation (KOSEF) grant funded by the Korean Government (MOST; no. R01-2007-000-11260-0) and National Research Foundation of Korea Grant funded by the Korean Government (2009-0075103).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Amino acid sequence comparison for each of six variable regions in Pvmsp-1 from Korean and worldwide isolates. Alignment of the sequence is based on a previous report (Putaporntip et al. 2002). Dashes indicate deletions. Short tandem repeats are in italic bold text with conserved sequences indicated in gray. The asterisk indicates a new type sequence identified in this study. (DOC 49 kb)
Supplementary Fig. 2
Amino acid sequence alignment of Pvmsp-1 conserved blocks from Korean isolates. The sequences were compared with Sal-1 and Belem type sequences. SKOR67, SKOR86, and SKOR83 sequences belong to SKOR-I, SKOR69 sequences to SKOR-II, SKOR0803 sequences to SKOR-III, SKOR0808 sequences to SKOR-IV, SKOR0804 sequences to SKOR-V, and SKOR0814 sequences to SKOR-VI allelic type. Single asterisk sequence that differs from those of the Sal-1 and Belem strains, colon dimorphic sequence, line deletion. Sal-1 and Belem type sequences are indicated by capital and italic text, respectively. Bold letters represent unique sequences from the Korean isolates. Amino acid positions shown above are after the Sal-1 sequence. CB conserved block. (DOC 41 kb)
Rights and permissions
About this article
Cite this article
Han, ET., Wang, Y., Lim, C.S. et al. Genetic diversity of the malaria vaccine candidate merozoite surface protein 1 gene of Plasmodium vivax field isolates in Republic of Korea. Parasitol Res 109, 1571–1576 (2011). https://doi.org/10.1007/s00436-011-2413-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-011-2413-5