Abstract
Major sperm proteins (MSPs) represent a protein family occurring in nematodes only. Identification of the 3′ and 5′ untranslated region (UTR) completed the so far partial msp complementary DNA sequences of the bovine lungworm Dictyocaulus viviparus. The full-length transcript contains sequence tracts consistent with the Kozak and polyadenylation consensus sequence. On genomic level, three full-length sequences differing in three nucleotides were determined containing a 65-bp phase zero intron. Conceptual translation inferred two MSP isoforms due to one substitution within the 126-amino acid polypeptide. Bioinformatic analysis predicted that bovine lungworm MSP folds into an immunoglobulin-like seven-stranded beta sandwich as known for Caenorhabditis elegans and Ascaris suum. Furthermore, bovine lungworm MSP is confidentially predicted to be N-terminal-acetylated and secreted via a non-classical pathway. Quantitative real-time polymerase chain reaction analysis using ten developmental lungworm stages showed that msp is transcribed mainly in adult male parasites and in some degree in hypobiotic L5. However, marginal msp transcription was detectable in all of the investigated developmental lungworm stages.
Similar content being viewed by others
Introduction
The family of major sperm proteins (commonly referred to as MSP) consists of closely related, small basic proteins playing an important role in reproductive processes. Until now, MSP orthologues have been found only within nematodes. In these, the protein is exclusively expressed in testicular tissues, but contributes to 15–17% of the total and 40% of the soluble protein in nematode sperm (Klass and Hirsh 1981; Nelson and Ward 1981; Ward et al. 1988). MSPs have a molecular weight of 14–15 kDa, an isoelectric point of about 8.0, and exist in solution as an extremely stable symmetrical dimer (Klass and Hirsh 1981; King et al. 1992; Haaf et al. 1996). In Caenorhabditis elegans, msp transcription starts in late primary spermatocytes and stops when the organelles necessary for protein synthesis remain with the residual body while the spermatids are budding (Ward and Klass 1982; Klass et al. 1982). These spermatids mature to spermatozoa, which, in contrast to other organisms than nematodes, are non-flagellated crawling cells. However, this amoeba-like pseudopodial movement is not actin-based, as the nematode sperm cytoskeleton is built from MSP assembled into filaments (Sepsenwol et al. 1989). Besides this cytoskeleton function, MSP acts as an extracellular signaling molecule that promotes oocyte meiotic maturation via an ephrin receptor and stimulates gonadal sheath cell contraction via antagonistic G protein signaling to facilitate ovulation (Miller et al. 2001; Govindan et al. 2006). As spermatozoa lack ribosomes, endoplasmic reticulum (ER), and Golgi for protein secretion, a vesicle-budding mechanism was identified in C. elegans delivering MSP to oocytes and sheath cells, the ovarian muscles (Kosinski et al. 2005).
Sequence entries of msp family member in the GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=Nucleotide) are available from only a few nematode species. From the free-living soil nematode C. elegans, 47 different messenger RNA (mRNA) as well as genomic sequences are deposited in this database. However, some of these genes are pseudogenes (cf. Wormbase, http://www.wormbase.org/). From mammalian parasitic nematodes, msp sequences in the GenBank nucleotide database were submitted from eight species only assigned to two of the five clades of the phylum Nematoda (Blaxter et al. 1998). Remarkably, most of the MSP sequences are based on genomic DNA. Of clade III, nucleotide data are available from Onchocerca volvulus (seven genomic sequences, five thereof partial), Mansonella ozzardi (17 partial genomic sequences), Brugia malayi (three partial mRNA sequences), Ascaris suum (one mRNA and one genomic sequence, each time the coding region only), and A. lumbricoides (one mRNA sequence). For the two latter organisms, each time, 11 sequences for MSP-domain-containing proteins are published beyond the msp sequences themselves. Clade V mammalian parasites with known msp nucleotide data are Oesophagostomum dentatum (two genomic sequences lacking the 5′ UTR and three mRNA sequences comprising the coding region only) as well as three lungworm species of the genus Dictyocaulus. However, even if 100 genomic msp sequence entries are deposited for the genus Dictyocaulus (72 sequence entries for D. viviparus, 18 sequence entries for D. eckerti, and ten sequence entries for D. capreolus), none of them is complete. Beyond these genomic sequences, one D. viviparus mRNA sequence (spanning the coding region only) is published.
In the GenBank EST database, the search for “major sperm protein” retrieves entries from additional mammalian parasitic nematodes, namely Trichuris vulpis, T. muris, and Trichinella spiralis from Nematoda Clade I; Litomosoides sigmodontis from Clade III; Strongyloides ratti and Parastrongyloides trichosuri from Clade IV, as well as Haemonchus contortus, Teladorsagia circumcincta, Ancylostoma caninum, A. ceylanicum, and Nippostrongylus brasiliensis from Clade V.
In terms of the very important functions of MSP for nematode reproduction, particularly its essential role for sperm motility as well as maturation and ovulation of the oocytes, which constitutes MSP as a promising candidate for drug and vaccine development, it is unexpected that sequences containing the 5′ and 3′ untranslated regions are only available in the databases for O. volvulus and A. lumbricoides.
In the present study, we completed the so far partial msp mRNA sequence of the bovine lungworm D. viviparus in 3′ and 5′ directions to obtain a full-length transcript. Furthermore, three full-length D. viviparus msp gene sequences were identified. The nucleotide and conceptually translated amino acid sequences were characterized on a molecular and bioinformatic level, respectively. A quantitative real-time polymerase chain reaction (PCR) assay was performed to investigate which lungworm stages transcribe msp. This transcriptional analysis was also used to answer the question if in D. viviparus female worms transcribe msp genes themselves for oocyte maturation and ovulation or if these features are mediated by male sperm MSP only.
Materials and methods
Parasite material for cDNA and genomic characterization
Adult parasites were produced in helminth-free Holstein–Friesian calves (5-month-old males) infected with 3300 L3. The calves were euthanized 28 days post-infection (p.i.) and the worms collected from the lungs as described by Wood et al. (1995). The worms were thoroughly washed five times in 200 ml saline solution (0.9%), transferred to sterile RNase free tubes, and frozen at −75°C.
Full-length cDNA
To obtain full-length msp complementary DNA (cDNA), RACE experiments were performed using the SMART™ RACE cDNA Amplification Kit (Clontech, Heidelberg, Germany) following the manufacturer’s recommendations. For this purpose, the PrimerSelect program of the Lasergene software (DNASTAR, version 5.06; GATC Biotech, Konstanz, Germany) was used to design gene-specific 3′ and 5′ RACE primers based on the known lungworm msp sequence fragment (accession no. S64873, Schnieder 1993). The full-length cDNA was generated by overlapping RACE sequences (Align Plus 5.0, Scientific and Educational Software, Cary, USA).
A PCR with primers spanning the whole sequence was carried out to verify the full-length cDNA sequence. Primer sequences for generating the full-length cDNA were: Dv msp for 5′-GATTAAGCGAAAGCTAAATCGG-3′ and Dv msp rev 5′-CGATTTTGTTAAGCATAGTGT-3′. PCR setup was as follows: 18 μl deionized H2O, 2.5 μl 10× buffer, 1.5 μl MgCl2 (25 mM), 1 μl deoxyribonucleotide triphosphates (2 mM each), 0.5 μl forward and reverse primer (50 μM each), respectively, and 0.125 μl HOT FIREpol® Polymerase (5 U/μl; Solis Biodyne, Tartu, Estonia) were added to 1 μl of template. PCR cycling (35 cycles) was performed using the following temperature profile: initial denaturing at 95°C for 15 min, denaturing at 95°C for 1 min, annealing primers at 60°C for 30 s, extending primers at 72°C for 1 min, and final extension at 72°C for 10 min. The band visible in gel electrophoresis (1% agarose gels) was cut out, ligated into pCR® 4-TOPO® vector followed by transformation of Escherichia coli One Shot® TOP 10 cells (TOPO TA Cloning® Kit for Sequencing; Invitrogen, Karlsruhe, Germany). Plasmid DNA was obtained using the NucleoSpin® Plasmid Kit (Macherey-Nagel, Dueren, Germany) following the manufacturer’s recommendations and sequenced at the SEQLAB Sequence laboratories (Göttingen, Germany).
Genomic characterization
Genomic DNA was isolated from ten pooled adult male and female worms using the NucleoSpin® Tissue Kit (Macherey-Nagel) following the manufacturer’s instructions with the exception that the genomic DNA was eluted twice in 50 μl of deionized H2O. The quality of isolated DNA was checked on a 1.0% agarose gel stained with GelStar® (Lonza, Wuppertal, Germany).
To obtain the msp genomic sequence, the full-length primer pair, PCR setup, and temperature profile from the above section were used. Cloning and sequencing was equally done as described above. Exon–intron boundaries were located by sequence comparison with the full-length cDNA generated in the above section using the program Align Plus 5.0 (Scientific and Educational Software).
Bioinformatic analysis
Prediction of protein structure, function, and posttranslational modifications was performed with the following internet service programs: PROFsec (http://www.aber.ac.uk/~phiwww/prof/) was used for prediction of the secondary structures helix, sheet, or loop based on multiple sequence alignments. Prediction results of D. viviparus MSP (accession nos. AAB27962, ABJ97284, and ABW37698) were compared to the known secondary structure of A. suum α-MSP (accession no. CAA63933) and C. elegans MSP-142 (accession no. NP_495144) described by Bullock et al. (1996) and Baker et al. (2002). Three-dimensional structure prediction of bovine lungworm MSP was done using the Protein Homology/analogY Recognition Engine version 0.2 (Phyre, http://www.sbg.bio.ic.ac.uk/phyre/). This program performs a profile–profile matching algorithm together with predicted secondary structure matching.
Identification and annotation of potential domains was achieved using the Simple Modular Architecture Research Tool (SMART) server (http://smart.embl-heidelberg.de/).
As in C. elegans MSP includes extracellular signaling molecule activity, we checked the bovine lungworm MSP for presence of a signal peptide cleavage site with the prediction program SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/). Furthermore, prediction of a potential non-classical and leaderless triggered secretion was performed using SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/). The possibility of N-terminal acetylation was analyzed using the NetAcet 1.0 server (http://www.cbs.dtu.dk/services/NetAcet/), predicting substrates of N-acetyltransferase A. Disulfide bonding state and cysteine connectivity prediction was performed with DISULFIND (http://disulfind.dsi.unifi.it/). Furthermore, prediction of N-glycosylation sites and phosphorylation sites of serine, threonine, and tyrosine was performed with the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) and the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos/), respectively.
Parasite material for quantitative real-time PCR
For quantitative real-time PCR, the following developmental stages were used: eggs; L1; L3; hypobiosis-induced L3 (L3i, chilled for 8 weeks at 4°C); male, female, and mixed gender L5; hypobiotic L5, as well as male and female adults of the strain HannoverDv2000. All stages were obtained using the methods described by Strube et al. (2008).
First-strand cDNA synthesis for quantitative real-time PCR
For poly(A)+ RNA isolation, the following amounts of the different lungworm stages were used: 75,000 eggs; 20,000 L1, L3, and L3i; 40 female and male L5; 20 females and 20 males as mixed gender L5 of both strains; 1,000 hypobiotic L5 as well as one female and male adult. Three separate setups of each lungworm stage were homogenized for 3 min in 200 μl extraction buffer (illustra™ QuickPrep Micro mRNA Purification Kit; GE Healthcare, Freiburg, Germany) using the TissueRuptor (Qiagen, Hilden, Germany) for subsequent poly(A)+ RNA isolation with the illustra™ QuickPrep Micro mRNA Purification Kit (GE Healthcare). The isolations were carried out following the manufacturer’s recommendations. Poly(A)+ RNA yield and purity was determined by measuring the absorbance at 260 and 280 nm using the NanoDrop® ND-1000 UV–Vis spectrophotometer (Peqlab, Erlangen, Germany). From each isolation, 300 ng poly(A)+ RNA was transcribed into cDNA by the use of Sprint™ PowerScript™ PrePrimed SingleShots with oligo(dT)18 primers (Clontech, Saint-Germain-en-Laye, France) according to the manufacturer’s instructions. The resulting first-strand cDNA was diluted 1:20 in tricine–EDTA buffer (10 mM tricine–KOH, pH 8.5; 1 mM EDTA).
Plasmid standards
Plasmid standards were used to compile standard curves for transcript copy number determination in the different lungworm stages. To generate plasmid standard inserts, msp and the housekeeping genes β-tubulin and elongation factor 1α (ef-1α) were amplified with gene-specific primers listed in Table 1.
Following ligation of the amplification products into the pCR® 4-TOPO® vector and transformation of E. coli One Shot® TOP 10 cells (TOPO TA Cloning® Kit for Sequencing; Invitrogen), the plasmids were isolated with the NucleoSpin® Plasmid Kit (Macherey-Nagel) following the manufacturer’s recommendations. To ensure sequence accuracy, the plasmids were sequenced at the SEQLAB Sequence laboratories (Göttingen, Germany). For the standards, tenfold serial plasmid dilutions ranging from 107 to 101 copies per sample were prepared.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed to determine msp transcription levels in different lungworm stages. According to Strube et al. (2008), the housekeeping genes β-tubulin and ef-1α were chosen to correct for variations of mRNA amounts and cDNA synthesis efficiency. Primers and TaqMan™ minor groove binder (MGB) probes were designed using the Primer Express software (Applied Biosystems, Darmstadt, Germany). Probes were purchased from Applied Biosystems and primers from Invitrogen. Primer sequences and corresponding TaqMan™ MGB probes are listed in Table 1. To avoid undesired detection of potentially contaminating genomic DNA, the selection criterion for a TaqMan™ MGB probe was to span a cDNA exon–exon junction.
First-strand cDNA preparations generated from three different mRNA isolations of the different lungworm stages (see “Parasite material for quantitative real-time PCR” and “First-strand cDNA synthesis for quantitative real-time PCR”) were used as template. The microtiter plates contained duplicates of each cDNA sample as well as serially diluted plasmid standards to generate standard curves and a no-template control. To ensure repeatability of the experiments, each run was replicated. qPCR was set up using the Brilliant® QPCR master mix (Stratagene, Heidelberg, Germany): 10.72 μl deionized H2O, 12.50 μl Brilliant buffer, 0.15 μl forward and reverse primers (50 μM each), respectively, 0.1 μl probe (50 μM), 0.38 μl diluted ROX as reference dye (1:500 dilution), and 1 μl first-strand cDNA template. Thermal cycling conditions were: 10 min at 95°C followed by 40 cycles of 20 s at 95°C, 20 s at 55°C, and 30 s at 72°C. Experiments and data analysis were performed using the Mx3005 Multiplex Quantitative PCR System (Stratagene). qPCR data mining was performed with the qBasePlus 1.0 software (Biogazelle, Zulte, Belgium).
Results
cDNA characterization of bovine lungworm msp
The full-length msp cDNA sequence (deposited in GenBank under accession no. EF012201) consists of 472 bp without poly(A)+ tail. The coding region is preceded by a 5′ untranslated region (UTR) of 29 bp and followed by 62 bp representing the 3′ UTR. The bases 426–431 within the 3′ UTR represent the polyadenylation signal 5′-AAUAAA-3′ leading to the covalent linkage of a poly(A)+ tail downstream nucleotide 472. The open reading frame has 381 bp encoding 126 deduced amino acids. The first ATG codon (bases 30–32) fulfills the requirement for a favorable context for initiation of translation, as there is a purine (A) in position −3 and the nucleotide G in position +4 (Kozak 1987). The encoded protein (accession no. AAB27962) has a theoretical molecular weight of 14.3 kDa and an isoelectric point of 7.99.
Organization of the D. viviparus msp gene
Amplification of genomic DNA to obtain the full-length msp gene sequence followed by sequencing of three clones resulted in three different sequences (deposited in GenBank under accession nos. DQ999999, EU140559, EU140560). These msp genes are divided into two exons and one intron with an entire length of 537 bp. The first 275 bp represents exon 1, which includes the 5′ UTR (bases 1–29) and the first 82 codons (bases 30–275) of the coding sequence. Exon 2 comprises the bases 341–357 (233 bp), whereby bases 341–475 represent the remaining 45 codons (including the termination codon) followed by the 3′ UTR. These two exons are flanking a phase zero intron (located between codons), which consists of 65 bp (bases 276–340). The bases GT are present at the 5′ splice site and AG at the 3′ splice site of the intron.
The analyzed msp genes showed sequence variations due to nucleotide substitutions in three positions. In nucleotide position 35, clone 1 (accession no. DQ999999) has an A instead of G present in the two other clones. However, this substitution has no consequence for the encoded amino acid (alanine), as it affects the wobble position in codon 2. In contrast, the nucleotide substitution of clone 2 (accession no. EU140559) of A for G in position 262, which represents the second base of codon 78, entails an amino acid substitution of glutamic acid for glycine. In consequence, the isoelectric point of the encoded protein (accession no. ABW37697) raises to 8.48 compared to 7.99 of the MSPs encoded by clones 1 and 3 (accession nos. ABJ97284 and ABW37698). Furthermore, clone 2 has a substitution from T to C in position 290, which is located within the intron.
Bioinformatic analysis
Protein structure and posttranslational modifications
The PROF secondary structure prediction system classified bovine lungworm MSP as an all-beta-sheet protein and identified nine sequence sections folding into beta strands. Figure 1 shows sequence and beta secondary structure comparison of D. viviparus MSP with α-MSP of A. suum and MSP-142 of C. elegans, which revealed identical folding of the polypeptide chains. Besides the beta strands, an alpha helix was predicted ranging from amino acid residues 100–110 within the polypeptide chain (cf. Fig. 1). Phyre prediction of a tertiary structure model of bovine lungworm MSP, which is illustrated in Fig. 2a,b, resulted in an estimated precision of 100%.
Sequence comparison between the amino acid residues and secondary beta sheet structure of D. viviparus MSP, A. suum α-MSP, and C. elegans MSP-142. Strands a1, b, c′1, and e form the three-stranded beta sheet, whereas strands a2, c, c′2, f, and g form the four-stranded beta sheet (further explanation in the text). This figure does not include the alpha helix structure comprising MSP amino acid residues 100–110 in D. viviparus and, accordingly, 101–111 in A. suum and C. elegans. Asterisk means identical amino acid residues in all sequences, colon means conserved substitutions, and dot means semi-conserved substitutions
The SMART server predicted with an expectation value of 1.00e−30 the Motile_Sperm, MSP domain (Pfam entry PF00635) within the bovine lungworm MSP ranging from amino acid residues 8 to 113. DISULFIND prediction of disulfide bonding and cysteine connectivity resulted in a not bonded protein state. The program SignalP did not locate a signal peptide cleavage site in D. viviparus MSP. However, SecretomeP classified it to be a non-classically secreted protein based on a score of 0.745 which exceeds the threshold of 0.5.
N-terminal acetylation was predicted by the NetAcet server for serine3 (score = 0.502), whereas alanine2 with a score of 0.464 failed to pass the threshold of 0.5. NetNGlyc predicted the asparagine35 with a potential of 0.5749 (threshold = 0.5) to be N-glycosylated. NetPhos identified as potential phosphorylation sites (threshold = 0.5) the amino acid residues serine37 (score = 0.861), threonine84 and 96 (scores = 0.916 and 0.960, respectively), as well as tyrosine23 (score = 0.881).
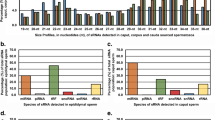
Quantitative real-time PCR
Plasmid dilution series ranging from 101 to 107 copies per microliter were used to compile qPCR standard curves for transcript copy number determination. The qBasePlus software calculated transcript amplification efficiencies based on the slope of the corresponding standard curve and squared correlation coefficients (R 2). These values were as follows: 68.4% (amplification efficiency), −4.418 (slope), and 0.983 (R2) for ef-1α; 78.9%, −3.957, and 0.995 for β-tubulin as well as 84.0%, −3.776, and 0.993 for msp. After ef-1α, β-tubulin, and msp raw qPCR data correction for their specific amplification efficiencies, the msp transcription profile was normalized to the reference genes ef-1α and β-tubulin. Relative quantification was used to determine the ratio between the quantity of msp transcripts in males as calibrator and the other developmental stages. As expected, msp transcription occurs mainly in adult male worms. The second highest transcription rate was observed in hypobiotic L5 with 23.836 copies in relation to 1,000 copies in adult males. In contrast, in “normal” L5, the relative amount of msp transcripts was 0.039 only. This relation of less than one copy compared to 1,000 adult male copies applies to the remaining populations as well. Table 2 lists relative quantities including standard error of the mean in more detail.
Discussion
Despite its essential role in nematode reproduction, almost all published msp nucleotide sequences lack at least the 5′ and 3′ UTR. The only parasite of human or veterinary importance for which full nucleotides data are available is so far O. volvulus.
In this study, we identified the full-length mRNA and genomic sequence of bovine lungworm msp followed by molecular characterization and analysis of its transcription pattern among ten developmental lungworm stages by quantitative real-time PCR.
The full-length msp mRNA sequence of D. viviparus spans 472 bp, of which 29 and 62 bp account for the 5′ and 3′ UTR, respectively. Within these, two important sequence sections were found: The purine A in position −3 preceding the first AUG codon is one of the two positions most critical for initiation of translation (Kozak 1986). The other most critical base in the consensus sequence for initiation is a G position +4, which was also present in the 5′ UTR of the bovine lungworm msp transcript. Furthermore, the polyadenylation consensus sequence 5′-AAUAAA-3′ was detected within the 3′ UTR.
Comparison of the full-length cDNA and genomic sequence revealed that in contrast to the published msp genes of the Clade V nematodes C. elegans and O. dentatum, which are intronless (Klass et al. 1984; Cottee et al. 2004), the D. viviparus msp gene contains a phase zero intron. This consists of 65 bp and follows the GT-AG rule (Breathnach et al. 1978). Interestingly, the so far partial msp gene sequences of D. eckerti and D. capreolus have shorter intron sequences of 59 bp only (Höglund et al. 2008). A single intron is also found in the msp genes of the clade III mammalian parasites A. suum, O. volvulus, and M. ozzardi (Hojas and Post 2000).
Three bovine lungworm genomic msp sequences differing in three of the overall 537 nucleotide positions were identified. Conceptual translation resulted in two MSP variants differing at one of the 126 residues. This substitution of glutamic acid for glycine in position 78 is non-isofunctional (Gindilis et al. 1998) and associated with a slightly more basic protein (pI of 8.48 vs. 7.99). In contrast, the two isoforms of A. suum, designated α- and β-MSP, differ at four out of 127 encoded amino acid residues (King et al. 1992), while in O. volvulus, the two isoforms differ at five out of 127 residues (Scott et al. 1989).
Secondary structure prediction of bovine lungworm MSP followed by comparison with A. suum α-MSP and C. elegans MSP-142 showed that the same stretches of amino acid residues form beta strands and an alpha helix, respectively (cf. Fig. 1). Therefore, it is safe to assume that D. viviparus MSP also folds into a single domain consisting of an immunoglobulin-type seven-stranded beta sandwich with opposing three-stranded and four-stranded beta sheets (Bullock et al. 1996; Baker et al. 2002). Indeed, this tertiary structure (cf. Fig. 2) was predicted by the Phyre program with an estimated precision of 100%. During tertiary folding, the C-terminal half of the first and the fourth strand (a2 and c′2) of A. suum and C. elegans MSP switch from one sheet to the other as a result of kinks introduced by cis-proline residues 13 and 57 (Bullock et al. 1996; Baker et al. 2002). This disruption of the strands is also predicted for bovine lungworm MSP having also proline residues directly following strands a1 and c′1. MSP polymerization to filaments or fibers starts with formation of homodimers which assemble into subfilaments. In this interaction, primarily residues 112–119 (Asp-Gly-Met-Arg-Arg-Lsy-Asn) are involved in terms of hydrogen bond formation between the g strands (Bullock et al. 1996, 1998). This amino acid stretch is conserved between various nematode species (Hojas and Post 2000) and also present in the two MSP variants of D. viviparus (cf. Fig. 1). MSP cysteine residues (positions 59, 74, and 94 in bovine lungworm MSP) provides the possibility of disulfide bonds not only between two monomers but also within them or both (Klass et al. 1984). However, the DISULFIND program predicted a not bonded state of the protein. This is in accordance to Haaf et al. (1996) who found that the two subunits in the MSP dimer were not covalently linked by a disulfide bond.
As expected, in both D. viviparus MSP variants, the Pfam domain Motile_Sperm, MSP (Pfam Entry PF00635), was identified by the SMART database. As reported by Miller et al. (2001) for C. elegans, the SignalP server predicts that bovine lungworm MSP lacks a signal peptide which is commonly known as a hallmark of secreted proteins. Nevertheless, MSP exhibits extracellular signaling function in addition to its intracellular cytoskeletal function for reproduction (Miller et al. 2001; Govindan et al. 2006). Due to the fact that nematode spermatids and spermatozoa are devoid of cellular components required for protein secretion, they release MSP by a non-classical secretory pathway identified as vesicle budding (Kosinski et al. 2005). Indeed, the SecretomeP server confidently predicted MSP secretion through a non-classical, i.e., not signal-peptide-triggered, mechanism. Since non-classical secretion is independent of the ER-Golgi network, the secreted proteins are never glycosylated even if they carry corresponding consensus sequences (Bendtsen et al. 2004), as it is the case with MSP which was found to contain a potential motif for N-glycosylation of asparagin35. Indeed, a non-glycosylated state was shown for C. elegans MSP (Burke and Ward 1983; Ward 1987). The same applies to potential MSP phosphorylation, which is predicted at four sites in D. viviparus but found to be absent in C. elegans MSP (Burke and Ward 1983; Ward 1987; Stewart et al. 1998). However, MSP might be phosphorylated transiently, like speculated by Haaf et al. (1998) in the case of MSP activation which is necessary for filament nucleation and elongation. A further predicted posttranslational modification of bovine lungworm MSP was N-terminal acetylation of serine3. This corresponds to A. suum MSP polypeptides for which mass spectroscopy yielded a mass consistent with initiator methionine cleavage and N-terminal acetylation (King et al. 1992), the two major types of protein modifications (Bradshaw et al. 1998; Polevoda and Sherman 2000). However, King et al. (1992) declare an acetylated alanine2 for both A. suum MSP isoforms, whereas the NetAcet server predicts serine3 of D. viviparus MSP to be acetylated (score = 0.502) instead of alanine2 whose score of 0.464 did not pass the threshold of 0.5. Acetylation regulates protein functions such as enzymatic activity, DNA binding, protein–protein interactions, peptide receptor recognition, and protein stability (Kouzarides 2000; Polevoda and Sherman 2002). Increased protein stability results in a longer protein half-life (Rudman et al. 1983; Martinez-Balbas et al. 2000), which might be the reason for N-terminal acetylation of MSP. Since neither spermatids nor mature sperms are capable of protein synthesis, MSP is expressed in spermatocytes only and stored in fibrous bodies and cytoplasm until required upon sperm activation (Ward and Klass 1982; Klass et al. 1982; Roberts et al. 1986).
Quantitative real-time PCR demonstrated that bovine lungworm msp is transcribed as a gender-specific transcript in adult male worms, whereas transcription in adult females is negligible. The ratio of 1,000 copies in males to 0.005 copies in females corresponds to a 200,000-fold higher transcription activity in males as expected from studies in other nematodes. In this context, no msp transcripts where detectable in females of C. elegans and O. dentatum (Ward 1987; Cottee et al. 2004), whereas the transcription rate in adult males of B. malayi is increased only 478-fold compared to female worms (Li et al. 2004). As described by Cottee et al. (2004) for O. dentatum, msp in D. viviparus is transcribed in a developmentally regulated manner as well. Besides adult males, the authors found O. dentatum msp to be transcribed in late L4, but not in L3 or early L4 stages. The bovine lungworm presents also a picture of developmental regulation: in the mixed L5 population isolated at day 15 p.i., 0.039 copies relative to 1,000 copies in adult males were determined compared to 23.836 relative copies in hypobiotic L5 isolated at day 27 p.i. This was unexpected, as hypobiosis is in general associated with postponed development. These results suggest that the hypobiotic slowdown affects not all developmental processes to the same extent. Therefore, hypobiotic L5 with a maximum of 5 mm in length (Pfeiffer 1976; Inderbitzin 1976) are shorter than normal L5 (about 10–20 mm at day 15 p.i., unpublished results), but their sexual development seems to be more advanced. In contrast to studies in other nematodes, we found msp transcription throughout all developmental stages of D. viviparus. However, transcription in other stages than hypobiotic L5 and adult male parasites are negligible, ranging from 0.005 relative copies in adult females to 0.319 relative copies in L3i (cf. Table 2). Since the msp translation product is a sperm component, posttranscriptional regulatory mechanisms most likely suppress protein translation of msp in early developmental stages or females. If normal or hypobiotic male L5 degrade msp transcripts or store them or even the translated protein for later use can only be speculation. Hence, it remains unclear if the msp transcript number in adult males is the consequence of partly accumulated transcripts or entirely transcribed in the adult stage.
In conclusion, the present study delivers insights into the organization of D. viviparus msp genes and their transcripts as well as the transcription levels throughout the bovine lungworm life cycle. However, further studies are needed to investigate if the three identified msp genes are equally or differentially transcribed or translated, respectively, as shown for A. suum. In this nematode, the β-MSP isoform is fivefold more abundant than α-MSP (King et al. 1992; Roberts and Stewart 1997). To clarify if msp transcripts are translated in earlier developmental stages than sexually mature males, immunohistochemical or other protein localizing studies would be desirable.
Since MSP is secreted and additionally absolutely essential for parasite reproduction, it represents a promising vaccine candidate. To determine its immunological and protective potential, vaccine trials against D. viviparus using recombinantly expressed MSP are underway.
References
Baker AM, Roberts TM, Stewart M (2002) 2.6 A resolution crystal structure of helices of the motile major sperm protein (MSP) of Caenorhabditis elegans. J Mol Biol 319:491–499
Bendtsen JD, Jensen LJ, Blom N, von HG, Brunak S (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17:349–356
Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM et al (1998) A molecular evolutionary framework for the phylum Nematoda. Nature 392:71–75
Bradshaw RA, Brickey WW, Walker KW (1998) N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem Sci 23:263–267
Breathnach R, Benoist C, O’Hare K, Gannon F, Chambon P (1978) Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon–intron boundaries. Proc Natl Acad Sci U S A 75:4853–4857
Bullock TL, Roberts TM, Stewart M (1996) 2.5 A resolution crystal structure of the motile major sperm protein (MSP) of Ascaris suum. J Mol Biol 263:284–296
Bullock TL, McCoy AJ, Kent HM, Roberts TM, Stewart M (1998) Structural basis for amoeboid motility in nematode sperm. Nat Struct Biol 5:184–189
Burke DJ, Ward S (1983) Identification of a large multigene family encoding the major sperm protein of Caenorhabditis elegans. J Mol Biol 171:1–29
Cottee PA, Nisbet AJ, Boag PR, Larsen M, Gasser RB (2004) Characterization of major sperm protein genes and their expression in Oesophagostomum dentatum (Nematoda: Strongylida). Parasitology 129:479–490
Gindilis V, Goltsman E, Verlinsky Y (1998) Evolutionary classification of homeodomains. J. Assist Reprod Genet 15:349–357
Govindan JA, Cheng H, Harris JE, Greenstein D (2006) Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C.elegans. Curr Biol 16:1257–1268
Haaf A, Butler PJ, Kent HM, Fearnley IM, Roberts TM, Neuhaus D, Stewart M (1996) The motile major sperm protein (MSP) from Ascaris suum is a symmetric dimer in solution. J Mol Biol 260:251–260
Haaf A, LeClaire L III, Roberts G, Kent HM, Roberts TM, Stewart M, Neuhaus D (1998) Solution structure of the motile major sperm protein (MSP) of Ascaris suum—evidence for two manganese binding sites and the possible role of divalent cations in filament formation. J Mol Biol 284:1611–1624
Höglund J, Engstrom A, Morrison DA, Mineur A, Mattsson JG (2008) Limited sequence variation in the major sperm protein 1 (MSP) gene within populations and species of the genus Dictyocaulus (Nematoda). Parasitol Res 103:11–20
Hojas RM, Post RJ (2000) Regional genetic variation in the major sperm protein genes of Onchocerca volvulus and Mansonella ozzardi (Nematoda: Filarioidea). Int J Parasitol 30:1459–1465
Inderbitzin F (1976) Experimentell erzeugte Entwicklungshemmung von Dictyocaulus viviparus des Rindes. PhD thesis, University of Zürich
King KL, Stewart M, Roberts TM, Seavy M (1992) Structure and macromolecular assembly of two isoforms of the major sperm protein (MSP) from the amoeboid sperm of the nematode, Ascaris suum. J Cell Sci 101(Pt 4):847–857
Klass MR, Hirsh D (1981) Sperm isolation and biochemical analysis of the major sperm protein from Caenorhabditis elegans. Dev Biol 84:299–312
Klass M, Dow B, Herndon M (1982) Cell-specific transcriptional regulation of the major sperm protein in Caenorhabditis elegans. Dev Biol 93:152–164
Klass MR, Kinsley S, Lopez LC (1984) Isolation and characterization of a sperm-specific gene family in the nematode Caenorhabditis elegans. Mol Cell Biol 4:529–537
Kosinski M, McDonald K, Schwartz J, Yamamoto I, Greenstein D (2005) C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132:3357–3369
Kouzarides T (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J 19:1176–1179
Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292
Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148
Li BW, Rush AC, Tan J, Weil GJ (2004) Quantitative analysis of gender-regulated transcripts in the filarial nematode Brugia malayi by real-time RT-PCR. Mol Biochem Parasitol 137:329–337
Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T (2000) Regulation of E2F1 activity by acetylation. EMBO J 19:662–671
Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D (2001) A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291:2144–2147
Nelson GA, Ward S (1981) Amoeboid motility and actin in Ascaris lumbricoides sperm. Exp Cell Res 131:149–160
Pfeiffer H (1976) Zur verzögerten Entwicklung des Rinderlungenwurmes, Dictyocaulus viviparus. Wien tierärztl Mschr 63:54–55
Polevoda B, Sherman F (2000) Nalpha-terminal acetylation of eukaryotic proteins. J Biol Chem 275:36479–36482
Polevoda B, Sherman F (2002) The diversity of acetylated proteins. Genome Biol 3:reviews0006
Roberts TM, Pavalko FM, Ward S (1986) Membrane and cytoplasmic proteins are transported in the same organelle complex during nematode spermatogenesis. J Cell Biol 102:1787–1796
Roberts TM, Stewart M (1997) Nematode sperm: amoeboid movement without actin. Trends Cell Biol 7:368–373
Rudman D, Hollins BM, Kutner MH, Moffitt SD, Lynn MJ (1983) Three types of alpha-melanocyte-stimulating hormone: bioactivities and half-lives. Am J Physiol 245:E47–E54
Schnieder T (1993) The diagnostic antigen encoded by gene fragment Dv3-14: a major sperm protein of Dictyocaulus viviparus. Int J Parasitol 23:383–389
Scott AL, Dinman J, Sussman DJ, Yenbutr P, Ward S (1989) Major sperm protein genes from Onchocerca volvulus. Mol Biochem Parasitol 36:119–126
Sepsenwol S, Ris H, Roberts TM (1989) A unique cytoskeleton associated with crawling in the amoeboid sperm of the nematode, Ascaris suum. J Cell Biol 108:55–66
Stewart M, Roberts TM, Italiano JE Jr, King KL, Hammel R, Parathasathy G, Bullock TL, McCoy AJ, Kent H, Haaf A, Neuhaus D (1998) Amoeboid motility without actin: insights into the molecular mechanism of locomotion using the major sperm protein (MSP) of nematodes. Biol Bull 194:342–343
Strube C, Buschbaum S, Wolken S, Schnieder T (2008) Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide isomerase transcription pattern in the bovine lungworm Dictyocaulus viviparus. Gene. doi:10.1016/j.gene.2008.08.001
Ward S (1987) Expression of sperm-specific genes during nematode spermatogenesis. Ann N Y Acad Sci 513:128–133
Ward S, Klass M (1982) The location of the major protein in Caenorhabditis elegans sperm and spermatocytes. Dev Biol 92:203–208
Ward S, Burke DJ, Sulston JE, Coulson AR, Albertson DG, Ammons D, Klass M, Hogan E (1988) Genomic organization of major sperm protein genes and pseudogenes in the nematode Caenorhabditis elegans. J Mol Biol 199:1–13
Wood IB, Amaral NK, Bairden K, Duncan JL, Kassai T, Malone JB Jr, Pankavich JA, Reinecke RK, Slocombe O, Taylor SM et al (1995) World Association for the Advancement of Veterinary Parasitology (W.A.A.V.P.) second edition of guidelines for evaluating the efficacy of anthelmintics in ruminants (bovine, ovine, caprine). Vet Parasitol 58:181–213
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide data reported in this paper have been submitted to the GenBank database under accession numbers EF012201, DQ999999, EU140559, and EU140560.
Rights and permissions
About this article
Cite this article
Strube, C., Buschbaum, S. & Schnieder, T. Molecular characterization and real-time PCR transcriptional analysis of Dictyocaulus viviparus major sperm proteins. Parasitol Res 104, 543–551 (2009). https://doi.org/10.1007/s00436-008-1228-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-008-1228-5