Abstract
Purpose
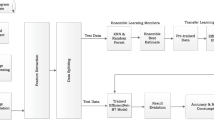
The classification of primary lung adenocarcinoma is complex and varied. Different subtypes of lung adenocarcinoma have different treatment methods and different prognosis. In this study, we collected 11 datasets comprising subtypes of lung cancer and proposed FL-STNet model to provide the assistance for improving clinical problems of pathologic classification in primary adenocarcinoma of lung.
Methods
Samples were collected from 360 patients diagnosed with lung adenocarcinoma and other subtypes of lung diseases. In addition, an auxiliary diagnosis algorithm based on Swin-Transformer, which used Focal Loss for function in training, was developed. Meanwhile, the diagnostic accuracy of the Swin-Transformer was compared to pathologists.
Results
The Swin-Transformer captures not only information in the overall tissue structure but also the local tissue details in the images of lung cancer pathology. Furthermore, training FL-STNet with the Focal Loss function can further balance the difference in the amount of data between different subtypes, improving recognition accuracy. The average classification accuracy, F1, and AUC of the proposed FL-STNet reached 85.71%, 86.57%, and 0.9903. The average accuracy of the FL-STNet was higher by 17% and 34%, respectively, than in the senior pathologist and junior pathologist group.
Conclusion
The first deep learning based on an 11-category classifier was developed for classifying lung adenocarcinoma subtypes based on WSI histopathology. Aiming at the deficiencies of the current CNN and Vit, FL-STNet model is proposed in this study by introducing Focal Loss and combining the advantages of Swin-Transformer model.







Similar content being viewed by others
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Aswolinskiy W, Tellez D, Raya G et al (2021) Neural image compression for non-small cell lung cancer subtype classification in H&E stained whole-slide images. In: Medical imaging 2021: digital pathology, vol 11603, p 1160304
Coudray N, Ocampo PS, Sakellaropoulos T et al (2018) Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 24(10):1559–1567
Dosovitskiy A, Beyer L, Kolesnikov A, et al (2020) An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929
Gertych A, Swiderska-Chadaj Z, Ma Z et al (2019) Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep 9(1):1–12
Graham S, Shaban M, Qaiser T, et al (2018) Classification of lung cancer histology images using patch-level summary statistics. In: SPIE Medical Imaging Symposium/6th Digital Pathology Conference
He K, Zhang X, Ren S, et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, pp 770–778
Hu WM, Li C, Li XY et al (2022) GasHisSDB: a new gastric histopathology image dataset for computer aided diagnosis of gastric cancer. Comput Biol Med 142:105207
Inamura K (2018) Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci 19(4):1259
Kazdal D, Rempel E, Oliveira C et al (2021) Conventional and semi-automatic histopathological analysis of tumor cell content for multigene sequencing of lung adenocarcinoma. Transl Lung Cancer Res 10(4):1666
Lin T-Y, Goyal P, Girshick R, et al (2017) Focal loss for dense object detection. In: Proceedings of the IEEE International Conference on computer vision, pp 2980–2988
Liu Z, Lin Y, Cao Y, et al (2021) Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on computer vision, pp 10012–10022
Maleki D, Tizhoosh H (2022) LILE: look in-depth before looking elsewhere—a dual attention network using transformers for cross-modal information retrieval in histopathology archives. In: Proceedings of the 5th international conference on medical imaging with deep learning, vol 172, pp 879–894
Meza R, Meernik C, Jeon J et al (2015) Lung cancer incidence trends by gender, race and histology in the United States, 1973–2010. PLoS ONE 10(3):e0121323
Murakami S, Ito H, Tsubokawa N et al (2015) Prognostic value of the new IASLC/ATS/ERS classification of clinical stage IA lung adenocarcinoma. Lung Cancer 90(2):199–204
Noguchi M, Morikawa A, Kawasaki M et al (1995) Small adenocarcinoma of the lung, histologic characteristics and prognosis. Cancer 75(12):2844–2852
Powers DM (2020) Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061
Russell PA, Wainer Z, Wright GM et al (2011) Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 6(9):1496–1504
Sarewitz S (2014) Subspecialization in community pathology practice. Arch Pathol Lab Med 138(7):871–872
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Tan M, Le Q (2019) Efficientnet: rethinking model scaling for convolutional neural networks. In: International Conference on machine learning, pp 6105–6114
Travis WD, Brambilla E, Nicholson AG et al (2015) The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10(9):1243–1260
Wang S, Yang DM, Rong R et al (2019a) Artificial intelligence in lung cancer pathology image analysis. Cancers 11(11):1673
Wang S, Wang T, Yang L et al (2019b) ConvPath: a software tool for lung adenocarcinoma digital pathological image analysis aided by a convolutional neural network. EBioMedicine 50:103–110
Wang C, Wu Y, Shao J et al (2020a) Clinicopathological variables influencing overall survival, recurrence and post-recurrence survival in resected stage I non-small-cell lung cancer. BMC Cancer 20(1):1–10
Wang S, Rong R, Yang DM et al (2020b) Computational staining of pathology images to study the tumor microenvironment in lung cancer. Can Res 80(10):2056–2066
Wei JW, Tafe LJ, Linnik YA et al (2019) Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep 9(1):1–8
Xia C, Dong X, Li H et al (2022) Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J 135(05):584–590
Yoshizawa A, Motoi N, Riely GJ et al (2011) Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 24(5):653–664
Yoshizawa A, Sumiyoshi S, Sonobe M et al (2013) Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol 8(1):52–61
Yu K-H, Wang F, Berry GJ et al (2020) Classifying non-small cell lung cancer types and transcriptomic subtypes using convolutional neural networks. J Am Med Inf Assoc 27(5):757–769
Zheng Y, Gindra RH, Green EJ et al (2022) A graph-transformer for whole slide image classification. IEEE Trans Med Imaging, 41(11):3003–3015
Acknowledgements
We thank the reviewers for their constructive comments.
Funding
The experimental fund was derived from Outstanding Talent Pool of Army Medical University (B-3257) and National Natural Science Foundation of China (52105265).
Author information
Authors and Affiliations
Contributions
(I) Conception and design:Yujun Wang, Furong Luo,Pan Huang and Hualiang Xiao participated in the conception of the experiment and the design of the technical route. (II) Provision of study materials or patients: The patient data collected in this study were all from hospitalized patients at Daping Hospital of Army Medical University. (III) Collection and assembly of data: Yujun Wang and Furong Luo were responsible for collecting digital image data and clinical information of patients in this study. (IV) Data analysis and interpretation: Yujun Wang and Furong Luo are responsible for the content analysis and interpretation of medical related research, while Xing Yang is responsible for the optimization of artificial intelligence algorithms and image training. (V) Manuscript writing: Yujun Wang, Furong Luo, Xing Yang, Qiushi Wang, Yunchun Sun, Sukun Tian, Peng Feng, Pan Huang, Hualiang Xiao. (VI) Final approval of manuscript: Yujun Wang, Furong Luo, Xing Yang, Qiushi Wang, Yunchun Sun, Sukun Tian, Peng Feng, Pan Huang, Hualiang Xiao.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Daping Hospital of Army Military Medical University (NO2022-216: the registration number of ethics board) and individual consent for this retrospective analysis was waived.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Luo, F., Yang, X. et al. The Swin-Transformer network based on focal loss is used to identify images of pathological subtypes of lung adenocarcinoma with high similarity and class imbalance. J Cancer Res Clin Oncol 149, 8581–8592 (2023). https://doi.org/10.1007/s00432-023-04795-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04795-y