Abstract
Purpose
The expression of eukaryotic translation initiation factor-2 subunit 3 (EIF2S3) in patients with non-small cell lung and colorectal cancer is lower than that in healthy individuals. However, the functions of EIF2S3 remain unclear, and its study in leukemia has not been reported. The article aims to explore the role of EIF2S3 in AML (acute myeloid leukemia) and its underlying mechanism.
Methods
Reverse transcription-quantitative PCR was performed to evaluate the expression levels of EIF2S3, and its association with patient prognosis was determined. Inducible HEL-EIF2S3 and HL-60-EIF2S3 cell lines were established by retrovirus infection. Cellular proliferation and the cell cycle were analyzed using Cell Counting Kit-8 and flow cytometric analyses. Tumorigenic ability was evaluated using xenograft nude mouse model. Gene expression profiles were analyzed in HL-60-EIF2S3 cells by next-generation sequencing, and WB analysis was performed to detect the expression of related proteins.
Results
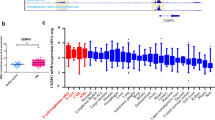
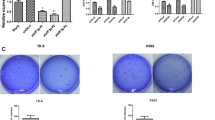
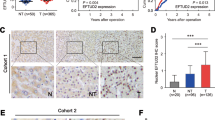
The expression of EIF2S3 in patients with AML was lower than that experiencing CR (P = 0.02). Furthermore, EIF2S3 overexpression inhibited cellular proliferation, halted G0/1 to S phase cell cycle progression, and inhibited tumorigenicity (P = 0.015). 479 differentially expressed genes were identified between HL60-EIF2S3 DOX (−) and HL60-EIF2S3 DOX ( +) cells via NGS and several of them involved in MAPK/ERK signaling pathway. The phosphorylation levels of ERK decreased when EIF2S3 was overexpressed (P < 0.050).
Conclusion
EIF2S3 overexpression may result in a decrease in cellular proliferation, cell cycle arrest, and tumorigenic inhibition via the MAPK/ERK signaling pathway in AML cells.






Similar content being viewed by others
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Arber DA, Orazi A, Hasserjian R et al (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127(20):2391–2405. https://doi.org/10.1182/blood-2016-08-733196
Borck G, Shin BS, Stiller B et al (2012) eIF2gamma mutation that disrupts eIF2 complex integrity links intellectual disability to impaired translation initiation. Mol Cell 48(4):641–646. https://doi.org/10.1016/j.molcel.2012.09.005
Bose P, Vachhani P, Cortes JE (2017) Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat Options Oncol 18(3):17. https://doi.org/10.1007/s11864-017-0456-2
Chang YT, Huang CS, Yao CT et al (2014) Gene expression profile of peripheral blood in colorectal cancer. World J Gastroenterol 20(39):14463–14471. https://doi.org/10.3748/wjg.v20.i39.14463
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132. https://doi.org/10.3322/caac.21338
Chian CF, Hwang YT, Terng HJ et al (2016) Panels of tumor-derived RNA markers in peripheral blood of patients with non-small cell lung cancer: their dependence on age, gender and clinical stages. Oncotarget 7(31):50582–50595. https://doi.org/10.18632/oncotarget.10558
Coombs CC, Tallman MS, Levine RL (2016) Molecular therapy for acute myeloid leukaemia. Nat Rev Clin Oncol 13(5):305–318. https://doi.org/10.1038/nrclinonc.2015.210
Dávila-Rodríguez MI, Cortés-Gutiérrez EI, Hernández-Valdés R, Guzmán-Cortés K, De León-Cantú RE, Cerda-Flores RM, Báez-De la Fuente E (2017) DNA damage in acute myeloid leukemia patients of Northern Mexico. Eur J Histochem 61(4):2851. https://doi.org/10.4081/ejh.2017.2851
Dohner H, Estey EH, Amadori S et al (2010) Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115(3):453–474. https://doi.org/10.1182/blood-2009-07-235358
Dohner H, Estey E, Grimwade D et al (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129(4):424–447
Grimwade D, Hills RK, Moorman AV et al (2010) Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116(3):354–365. https://doi.org/10.1182/blood-2009-11-254441
Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y, Hu LL (2020) ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med 19(3):1997–2007. https://doi.org/10.3892/etm.2020.8454
Hinnebusch AG (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75(3):434–467. https://doi.org/10.1128/MMBR.00008-11 (first page of table of contents)
Hinnebusch AG (2014) The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812. https://doi.org/10.1146/annurev-biochem-060713-035802
Hinnebusch AG, Lorsch JR (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a011544
Lavoie H, Therrien M (2015) Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol 16(5):281–298. https://doi.org/10.1038/nrm3979
Lee JT Jr, McCubrey JA (2002) The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia 16(4):486–507. https://doi.org/10.1038/sj.leu.2402460
Lord CJ, Ashworth A (2012) The DNA damage response and cancer therapy. Nature 481(7381):287–294. https://doi.org/10.1038/nature10760
Meloche S, Pouyssegur J (2007) The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 26(22):3227–3239. https://doi.org/10.1038/sj.onc.1210414
Moortgat S, Desir J, Benoit V et al (2016) Two novel EIF2S3 mutations associated with syndromic intellectual disability with severe microcephaly, growth retardation, and epilepsy. Am J Med Genet A 170(11):2927–2933. https://doi.org/10.1002/ajmg.a.37792
Narayanan D, Weinberg OK (2020) How I investigate acute myeloid leukemia. Int J Lab Hematol 42(1):3–15. https://doi.org/10.1111/ijlh.13135
Pennisi R, Albanesi J, Ascenzi P, Nervi C, di Masi A (2018) Are DNA damage response kinases a target for the differentiation treatment of acute myeloid leukemia? IUBMB Life 70(11):1057–1066. https://doi.org/10.1002/iub.1918
Saez-Rodriguez J, MacNamara A, Cook S (2015) Modeling signaling networks to advance new cancer therapies. Annu Rev Biomed Eng 17:143–163. https://doi.org/10.1146/annurev-bioeng-071813-104927
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70(1):7–30. https://doi.org/10.3322/caac.21590
Skopkova M, Hennig F, Shin BS et al (2017) EIF2S3 mutations associated with severe X-linked intellectual disability syndrome MEHMO. Hum Mutat 38(4):409–425. https://doi.org/10.1002/humu.23170 (Epub 2017 Jan 23)
Stanik J, Skopkova M, Stanikova D et al (2018) Neonatal hypoglycemia, early-onset diabetes and hypopituitarism due to the mutation in EIF2S3 gene causing MEHMO syndrome. Physiol Res 67(2):331–337. https://doi.org/10.33549/physiolres.933689 (Epub 2018 Jan 5)
Su Y, Li X, Ma J et al (2018) Targeting PI3K, mTOR, ERK, and Bcl-2 signaling network shows superior antileukemic activity against AML ex vivo. Biochem Pharmacol 148:13–26. https://doi.org/10.1016/j.bcp.2017.11.022
Vasjari L, Bresan S, Biskup C, Pai G, Rubio I (2019) Ras signals principally via Erk in G1 but cooperates with PI3K/Akt for Cyclin D induction and S-phase entry. Cell Cycle 18(2):204–225. https://doi.org/10.1080/15384101.2018.1560205
Wong RS (2011) Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res 30(1):87. https://doi.org/10.1186/1756-9966-30-87
Yamamoto T, Ebisuya M, Ashida F, Okamoto K, Yonehara S, Nishida E (2006) Continuous ERK activation downregulates antiproliferative genes throughout G1 phase to allow cell-cycle progression. Curr Biol 16(12):1171–1182. https://doi.org/10.1016/j.cub.2006.04.044
Young-Baird SK, Shin BS, Dever TE (2019) MEHMO syndrome mutation EIF2S3-I259M impairs initiator Met-tRNAiMet binding to eukaryotic translation initiation factor eIF2. Nucleic Acids Res 47(2):855–867. https://doi.org/10.1093/nar/gky1213
Funding
The present study was supported by the National Natural Science Foundation of China (Grant no. 81672661, 81870113) and the Guangzhou Science and Technology Project (grant no. 201804010199).
Author information
Authors and Affiliations
Contributions
LX, YZ, and ZH conceived the study idea, supervised the project and designed the experiments; JL and SC performed the experiments and wrote the manuscript; LL, HT, BL, YC, and XZ collected the samples, performed the experiments, and analyzed the data.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The present study was approved by the ethics committee of the First Affiliated Hospital of Guangzhou Medical University. All patients provided written informed consent for the use of their clinical specimens. Animals were purchased from the Animal experimental center, Shanghai, China, and the protocol was approved by the Institutional Animal Care and Use Committee of Guangzhou Medical University. All mouse procedures were performed in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of Guangzhou Medical University. Animal experiments were performed at Guangzhou Medical University and all surgeries were performed under sodium pentobarbital anesthesia; all efforts were made to minimize animal suffering.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
432_2021_3712_MOESM1_ESM.tif
Supplementary file1 Fig. S1 EIF2S3 mRNA expression level in AML patients of TCGA based on classification of clinical characteristics. A The difference of EIF2S3 mRNA expression based on FBA types. EIF2S3 expression in M3 was lower than M0 (P=0.019), M1 (P=0.001), M2 (P=0.004) and M4 (P=0.022) patients. EIF2S3 expression in M5 patients was lower than M0 (P=0.012), M1 (P<0.001), M2 (P<0.001) and M4 (P=0.003) patients. There were no significant differences between other types. B The difference of EIF2S3 mRNA expression based on gender (P=0.015). C The difference of EIF2S3 mRNA expression based on age. EIF2S3 expression in 21-40 years-old patient was higher than 81-100 years-old patients (P=0.048). There were no significant differences in other age groups. D The difference of EIF2S3 mRNA expression based on races (P>0.05) (TIF 634 KB)
432_2021_3712_MOESM2_ESM.tif
Supplementary file2 Fig. S2 EIF2S3 mRNA expression level in AML patients of TCGA based on classification of genetic abnormality. A The difference of EIF2S3 mRNA expression based on FLT3 mutation or not (P>0.05). B The difference of EIF2S3 mRNA expression based on PML/RAR fusion or not (P>0.05). C The difference of EIF2S3 mRNA expression based on RAS activation or not (P>0.05) (TIF 14657 KB)
432_2021_3712_MOESM3_ESM.tif
Supplementary file3 Fig. S3 EIF2S3 mRNA expression levels in patients with primary AML (PB) and healthy controls. Comparing with health controls, EIF2S3 expression was downregulated in primary AML patients (PB) (P =0.009). PB, peripheral blood (TIF 81 KB)
432_2021_3712_MOESM4_ESM.tif
Supplementary file4 Fig. S4 Identify the DEGs in HL60-EIF2S3 DOX (-) and HL60-EIF2S3 DOX (+) by NGS. A Using P<0.05 and |log2FC|≥2 as cut-0ff criteria, the volcano plot of DEGs showed there were totally 479 DEGs, including 271 upregulated genes (red dots) and 208 downregulated genes (blue dots). B In the hot plot of top 30 DEGs, red represents upregulation and blue represent downregulation. The closer the color was to the ends of the color column, the greater the difference (TIF 9749 KB)
432_2021_3712_MOESM5_ESM.tif
Supplementary file5 Fig. S5 GO enrichment analysis in HL60-EIF2S3 DOX (-) and HL60-EIF2S3 DOX (+). The top 30 of GO enrichment described relevant biological processes of DEGs. The color of the dots represents the significance of q value, and the size of the dots represents the number of DEGs involved in the GO domain (TIF 24273 KB)
432_2021_3712_MOESM6_ESM.tif
Supplementary file6 Fig. S6 KEGG enrichment analysis in HL60-EIF2S3 DOX (-) and HL60-EIF2S3 DOX (+). The top 30 of pathway enrichment showed that DEGs primarily enriched in MAPK pathway. The color of the dots represents the significance of q value, and the size of the dots represents the number of DEGs involved in the different pathways (TIF 11516 KB)
432_2021_3712_MOESM7_ESM.tif
Supplementary file7 Fig. S7 RT-qPCR showed the relative mRNA expression level of 13 DEGs in MAPK signaling pathway. *P < 0.05, **P < 0.01, and ***P < 0.001 (TIF 473 KB)
Rights and permissions
About this article
Cite this article
Lu, J., Chen, S., Tan, H. et al. Eukaryotic initiation factor-2, gamma subunit, suppresses proliferation and regulates the cell cycle via the MAPK/ERK signaling pathway in acute myeloid leukemia. J Cancer Res Clin Oncol 147, 3157–3168 (2021). https://doi.org/10.1007/s00432-021-03712-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-021-03712-5