Abstract
Purpose
Microvascular invasion (MVI) is a critical determinant of the early recurrence and poor prognosis of patients with hepatocellular carcinoma (HCC). Prediction of MVI status is clinically significant for the decision of treatment strategies and the assessment of patient’s prognosis. A deep learning (DL) model was developed to predict the MVI status and grade in HCC patients based on preoperative dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and clinical parameters.
Methods
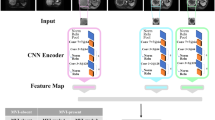
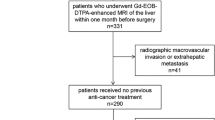
HCC patients with pathologically confirmed MVI status from January to December 2016 were enrolled and preoperative DCE-MRI of these patients were collected in this study. Then they were randomly divided into the training and testing cohorts. A DL model with eight conventional neural network (CNN) branches for eight MRI sequences was built to predict the presence of MVI, and further combined with clinical parameters for better prediction.
Results
Among 601 HCC patients, 376 patients were pathologically MVI absent, and 225 patients were MVI present. To predict the presence of MVI, the DL model based only on images achieved an area under curve (AUC) of 0.915 in the testing cohort as compared to the radiomics model with an AUC of 0.731. The DL combined with clinical parameters (DLC) model yielded the best predictive performance with an AUC of 0.931. For the MVI-grade stratification, the DLC models achieved an overall accuracy of 0.793. Survival analysis demonstrated that the patients with DLC-predicted MVI status were associated with the poor overall survival (OS) and recurrence-free survival (RFS). Further investigation showed that hepatectomy with the wide resection margin contributes to better OS and RFS in the DLC-predicted MVI present patients.
Conclusion
The proposed DLC model can provide a non-invasive approach to evaluate MVI before surgery, which can help surgeons make decisions of surgical strategies and assess patient’s prognosis.





Similar content being viewed by others
Data availability
Data are available in the Article and Supplementary Information. All other data can be provided upon reasonable request to the corresponding authors.
Code availability
The code of the proposed method can be provided upon reasonable request to the corresponding authors.
Abbreviations
- MVI:
-
Microvascular invasion
- HCC:
-
Hepatocellular carcinoma
- DL:
-
Deep learning
- DCE-MRI:
-
Dynamic contrast-enhanced magnetic resonance imaging
- CNN:
-
Conventional neural network
- AUC:
-
Area under curve
- DLC:
-
Deep learning combined with clinical parameters
- OS:
-
Overall survival
- RFS:
-
Recurrence-free survival
- RFA:
-
Radiofrequency ablation
- AFP:
-
Alpha fetoprotein
- PIVKA-II:
-
Prothrombin induced by vitamin K absence-II
- CT:
-
Computed tomography
- MRI:
-
Magnetic resonance imaging
- PLR:
-
Platelet-lymphocyte ratio
- NLR:
-
Neutrophil-lymphocyte ratio
- LMR:
-
Lymphocyte-to-monocyte ratio
- PNI:
-
Prognostic nutritional index
- APRI:
-
Aspartate aminotransferase-to-platelet ratio index
- ANRI:
-
Aspartate aminotransferase-to-neutrophil ratio index
- ALR:
-
Aspartate aminotransferase-lymphocyte ratio
- T2WI:
-
Turbo spin-echo T2-weighted
- DWI:
-
Diffusion-weighted imaging
- ADC:
-
Apparent diffusion coefficient
- 3D:
-
Three-dimensional
- VOI:
-
Volume of interest
- FC:
-
Fully connected
- AIC:
-
Akaike information criterion
- ROC:
-
Receiver operating characteristic
- INR:
-
International normalized ratio
- SD:
-
Standard deviation
- IQR:
-
Interquartile range
- CI:
-
Confidence interval
- NCCN:
-
National Comprehensive Cancer Network
- AASLD:
-
American Association for the Study of Liver Diseases
References
Aerts HJWL (2016) The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol 2:1636–1642. https://doi.org/10.1001/jamaoncol.2016.2631
Ardila D et al (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 25:954–961. https://doi.org/10.1038/s41591-019-0447-x
Cong WM, Bu H, Chen J, Dong H, Zhu YY, Feng LH, Chen J (2016) Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J Gastroenterol 22:9279–9287. https://doi.org/10.3748/wjg.v22.i42.9279
Coudray N et al (2018) Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24:1559–1567. https://doi.org/10.1038/s41591-018-0177-5
Han HS, Shehta A, Ahn S, Yoon YS, Cho JY, Choi Y (2015) Laparoscopic versus open liver resection for hepatocellular carcinoma: case-matched study with propensity score matching. J Hepatol 63:643–650. https://doi.org/10.1016/j.jhep.2015.04.005
Han J et al (2019) The impact of resection margin and microvascular invasion on long-term prognosis after curative resection of hepatocellular carcinoma: a multi-institutional study. HPB (Oxford) 21:962–971. https://doi.org/10.1016/j.hpb.2018.11.005
Hasegawa K et al (2013) Comparison of resection and ablation for hepatocellular carcinoma: a cohort study based on a Japanese nationwide survey. J Hepatol 58:724–729. https://doi.org/10.1016/j.jhep.2012.11.009
Imai K et al (2018) Microvascular invasion in small-sized hepatocellular carcinoma: significance for outcomes following hepatectomy and radiofrequency ablation. Anticancer Res 38:1053–1060. https://doi.org/10.21873/anticanres.12322
Ishizawa T et al (2008) Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology 134:1908–1916. https://doi.org/10.1053/j.gastro.2008.02.091
Jiang YQ et al (2021) Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J Cancer Res Clin Oncol 147:821–833. https://doi.org/10.1007/s00432-020-03366-9
Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY (2019) Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res 25:3847–3855. https://doi.org/10.1158/1078-0432.Ccr-18-2861
Lee S, Kim SH, Lee JE, Sinn DH, Park CK (2017) Preoperative gadoxetic acid-enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J Hepatol 67:526–534. https://doi.org/10.1016/j.jhep.2017.04.024
Lee S et al (2021) Effect of microvascular invasion risk on early recurrence of hepatocellular carcinoma after surgery and radiofrequency ablation. Ann Surg 273:564–571. https://doi.org/10.1097/sla.0000000000003268
Lim KC et al (2011) Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg 254:108–113. https://doi.org/10.1097/SLA.0b013e31821ad884
Liu Z et al (2019) The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 9:1303–1322. https://doi.org/10.7150/thno.30309
Liu Y et al (2020) A deep learning system for differential diagnosis of skin diseases. Nat Med 26:900–908. https://doi.org/10.1038/s41591-020-0842-3
Ma X et al (2019) Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol 29:3595–3605. https://doi.org/10.1007/s00330-018-5985-y
Marrero JA et al (2018) Diagnosis, staging, and management of hepatocellular carcinoma practice guidance by the American association for the study of liver diseases. Hepatology 68:723–750. https://doi.org/10.1002/hep.29913
Mazzaferro V et al (2009a) Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol 10:35–43. https://doi.org/10.1016/S1470-2045(08)70284-5
NCCN. National Comprehensive Cancer Network Clinical Practice Guidelines in oncology: hepatobiliary cancers, version 2.2019. https://www.nccn.org/professionals/physician_gls/pdf/hepatobiliary.pdf. Accessed Apr 2019
Nitta H et al (2019) Prognostic value and prediction of extratumoral microvascular invasion for hepatocellular carcinoma. Ann Surg Oncol 26:2568–2576. https://doi.org/10.1245/s10434-019-07365-0
Park CM (2019) Can artificial intelligence fix the reproducibility problem of radiomics? Radiology 292:374–375. https://doi.org/10.1148/radiol.2019191154
Poté N et al (2015) Performance of PIVKA-II for early hepatocellular carcinoma diagnosis and prediction of microvascular invasion. J Hepatol 62:848–854. https://doi.org/10.1016/j.jhep.2014.11.005
Price WN (2018) Big data and black-box medical algorithms. Sci Transl Med 10:eaao5333. https://doi.org/10.1126/scitranslmed.aao5333
Reginelli A et al (2018) Can microvascular invasion in hepatocellular carcinoma be predicted by diagnostic imaging? A critical review. Future Oncol 14:2985–2994. https://doi.org/10.2217/fon-2018-0175
Roayaie S et al (2009) A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology 137:850–855. https://doi.org/10.1053/j.gastro.2009.06.003
Roberts LR et al (2018) Imaging for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Hepatology 67:401–421. https://doi.org/10.1002/hep.29487
Rodríguez-Perálvarez M, Luong TV, Andreana L, Meyer T, Dhillon AP, Burroughs AK (2013) A systematic review of microvascular invasion in hepatocellular carcinoma: diagnostic and prognostic variability. Ann Surg Oncol 20:325–339. https://doi.org/10.1245/s10434-012-2513-1
Shindoh J et al (2013) Risk factors of post-operative recurrence and adequate surgical approach to improve long-term outcomes of hepatocellular carcinoma. HPB (Oxford) 15:31–39. https://doi.org/10.1111/j.1477-2574.2012.00552.x
Sun JJ et al (2016) Postoperative adjuvant transcatheter arterial chemoembolization after R0 hepatectomy improves outcomes of patients who have hepatocellular carcinoma with microvascular invasion. Ann Surg Oncol 23:1344–1351. https://doi.org/10.1245/s10434-015-5008-z
Ting DSW et al (2017) Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 318:2211–2223. https://doi.org/10.1001/jama.2017.18152
Tsilimigras DI et al (2020) Effect of surgical margin width on patterns of recurrence among patients undergoing R0 hepatectomy for T1 hepatocellular carcinoma: an international multi-institutional analysis. J Gastrointest Surg 24:1552–1560. https://doi.org/10.1007/s11605-019-04275-0
Villanueva A (2019) Hepatocellular carcinoma. N Engl J Med 380:1450–1462. https://doi.org/10.1056/NEJMra1713263
Vitale A et al (2014) Is resectable hepatocellular carcinoma a contraindication to liver transplantation? A novel decision model based on “number of patients needed to transplant” as measure of transplant benefit. J Hepatol 60:1165–1171. https://doi.org/10.1016/j.jhep.2014.01.022
Wang P et al (2018) Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng 2:741–748. https://doi.org/10.1038/s41551-018-0301-3
Wang S et al (2019) Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 53:1800986. https://doi.org/10.1183/13993003.00986-2018
Wang L, Jin YX, Ji YZ, Mu Y, Zhang SC, Pan SY (2020a) Development and validation of a prediction model for microvascular invasion in hepatocellular carcinoma. World J Gastroenterol 26:1647–1659. https://doi.org/10.3748/wjg.v26.i14.1647
Wang L et al (2020b) Postoperative adjuvant treatment strategy for hepatocellular carcinoma with microvascular invasion: a non-randomized interventional clinical study. BMC Cancer 20:614. https://doi.org/10.1186/s12885-020-07087-7
Xu X et al (2019) Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol 70:1133–1144. https://doi.org/10.1016/j.jhep.2019.02.023
Zeng F, Chen B, Zeng J, Wang Z, Xiao L, Deng G (2019) Preoperative neutrophil-lymphocyte ratio predicts the risk of microvascular invasion in hepatocellular carcinoma: a meta-analysis. Int J Biol Markers 34:213–220. https://doi.org/10.1177/1724600819874487
Zhang XP et al (2019) Postoperative adjuvant sorafenib improves survival outcomes in hepatocellular carcinoma patients with microvascular invasion after R0 liver resection: a propensity score matching analysis. HPB (Oxford) 21:1687–1696. https://doi.org/10.1016/j.hpb.2019.04.014
Zhang X et al (2020b) Contrast-enhanced CT radiomics for preoperative evaluation of microvascular invasion in hepatocellular carcinoma: a two-center study. Clin Transl Med 10:e111. https://doi.org/10.1002/ctm2.111
Zhang H et al (2020a) Predictive value of gamma-glutamyl transpeptidase to lymphocyte count ratio in hepatocellular carcinoma patients with microvascular invasion. BMC Cancer 20:132. https://doi.org/10.1186/s12885-020-6628-7
Zheng J et al (2017) Utility of serum inflammatory markers for predicting microvascular invasion and survival for patients with hepatocellular carcinoma. Ann Surg Oncol 24:3706–3714. https://doi.org/10.1245/s10434-017-6060-7
Zhou J et al (2018) Guidelines for diagnosis and treatment of primary liver cancer in China (2017 Edition). Liver Cancer 7:235–260. https://doi.org/10.1159/000488035
Zhu Y et al (2018) A new laboratory-based algorithm to predict microvascular invasion and survival in patients with hepatocellular carcinoma. Int J Surg 57:45–53. https://doi.org/10.1016/j.ijsu.2018.07.011
Acknowledgement
We acknowledged the Department of Pathology of Zhongshan Hospital of Fudan University for assistance with pathologic diagnosis.
Funding
This work was supported by National Key Research and Development Program of China (Grant 82090054) and National Natural Science Foundation of China (Grant 81572367 and 81772556) to Xiaoying Wang, Shanghai Science and Technology Innovation Action Plan (Grant 19511121302) to Manning Wang, National Key Research and Development Program of China (Grant 2017YFC0108804) to Shengxiang Rao and Shanghai Sailing Program (Grant 19YF1408100) to Wentao Wang.
Author information
Authors and Affiliations
Contributions
WXY, WMN, RSX: Guarantors of integrity of entire study; WXY: Concepts and design; All authors: Administrative support, Data acquisition (including radiological and clinical data); SDJ, WWT, RSX, WXY: Radiological images delineation; WYY, WMN: Development of methodology; SDJ, WYY, LMZ: Statistical analysis; SDJ, WYY: Manuscript drafting or revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors of this manuscript declare no conflict of interest.
Ethical approval
This retrospective study was approved by IRB and the requirement for written informed consent was waived by IRB.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Song, D., Wang, Y., Wang, W. et al. Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J Cancer Res Clin Oncol 147, 3757–3767 (2021). https://doi.org/10.1007/s00432-021-03617-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-021-03617-3