Abstract
Purpose
Non-neuroendocrine neoplasms of the appendix are a phenotypically heterogeneous group of lesions; a comprehensive molecular characterization of these tumors is still lacking.
Methods
A total of 52 samples taken from 49 patients was evaluated: 18 sessile serrated lesions (SSL; 3 with dysplasia), 2 high-grade tubular adenomas, 1 tubulo-villous adenoma,1 hyperplastic polyp, 18 low-grade appendiceal mucinous neoplasms (LAMN), 3 high-grade appendiceal mucinous neoplasms (HAMN) and 9 mucinous adenocarcinomas. Hotspot mutational profiling of the RNF43, SMAD4, KRAS, NRAS, BRAF and PIK3CA genes was performed. Expression of p53, MLH1, PMS2, MSH2, and MSH6 was evaluated by immunohistochemistry.
Results
KRAS was the most frequently mutated gene (53.9% of cases), followed by RNF43 (15.4%), and BRAF (13.5%). In particular: KRAS was mutated in 44.4% of adenocarcinomas, 66.7% of HAMNs, 61.1% of LAMNs, 53.3% of SSL without dysplasia and in 66.7% of SSL with dysplasia; RNF43 was mutated in 33.3% of adenocarcinomas, 66.7% of HAMNs, 11.1% of LAMNs and in 6.7% of SSL without dysplasia; BRAF was mutated in 11.1% of adenocarcinomas, 26.7% of SSL without dysplasia and in 5.6% of LAMNs. Only a case of high-grade tubular adenoma showed mismatch repair deficiency, while immunohistochemical expression of p53 was altered in 21.1% of cases.
Conclusions
The histological phenotypic similarities between appendicular mucinous lesions and serrated colon lesions do not reflect a similar genetic landscape. Mismatch repair deficiency is a rare event during appendiceal mucinous carcinogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary tumours of the appendix are reported in less than 1% of appendectomy specimens (Connor et al. 1998; Smeenk et al. 2008; WHO Classification of Tumours Editorial Board 2019).
Among the others, the appendicular epithelial non-neuroendocrine neoplasms are a heterogeneous group of lesions, that the Peritoneal Surface Oncology Group International (PSOGI) have proposed to classify in two main groups: non-invasive and invasive neoplasms (Carr et al. 2016). Sessile serrated lesions with or without dysplasia (SSLd/SSL), conventional adenoma resembling colorectal type, low-grade appendiceal mucinous neoplasm (LAMN), and high-grade appendiceal mucinous neoplasm (HAMN) belong to the non-invasive group; while mucinous adenocarcinoma, mucinous adenocarcinoma with signet ring cells, signet ring cell carcinoma and non-mucinous carcinoma are considered invasive neoplasms (Carr et al. 2016). All these neoplasms have the common potential to cause pseudomyxoma peritonei (PMP). PMP is a clinical entity characterized by grossly evident, diffuse, intra-abdominal mucinous ascites and by peritoneal implants (Carr et al. 2016).
Early-stage appendicular neoplasms can be treated with surgery and long-term benefits have also been achieved with cytoreductive surgery and heated intraperitoneal chemotherapy (HIPEC) (Reghunathan et al. 2018). On the contrary, neither a standard of care nor specific targeted therapies currently exist for patients with advanced or unresectable tumours. Despite the lack of consensus, chemotherapeutic regimens approved for colorectal cancer (CRC) are most commonly used for advanced cases (Tokunaga et al. 2019; Pietrantonio et al. 2014).
Disseminated appendiceal mucinous neoplasms exhibit a spectrum of clinical behaviours, ranging from slow-growing tumours with considerable risk of recurrence and eventual death, to highly aggressive neoplasms with a high risk of early death (Carr et al. 2012; Davison et al. 2014).
Although in the last few years an increasing attention has been paid to improve the classification and definition of appendicular neoplasms, a deeper molecular characterization of these tumors is still lacking.
Materials and methods
Patients and tumor samples
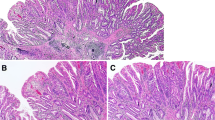
A total of 52 appendiceal lesions from 49 Caucasian patients (F/M = 26/24; age 71.95 ± 13.3) were collected from 2011 to 2018 in the Department of Surgical Pathology, University of Padua Medical Centre. Pathology reports and hospital charts were reviewed to collect the following information: type of surgical procedure, age at presentation, gender, and anatomic extent of tumour at diagnosis. All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Declaration of Helsinki. The 52 appendiceal lesions were jointly re-evaluated by three gastrointestinal pathologists according to 2019 WHO classification of tumours of the digestive system (WHO Classification of Tumours Editorial Board 2019) and classified as: 18 serrated lesions (3 with dysplasia), 2 high-grade tubular adenomas, 1 tubulo-villous adenoma, 1 hyperplastic polyp, 18 low-grade appendiceal mucinous neoplasms (LAMN), 3 high-grade appendiceal mucinous neoplasms (HAMN) and 9 mucinous adenocarcinomas (no signet-ring cell adenocarcinoma was considered; Fig. 1).
Representative examples of the considered series of appendiceal mucinous neoplasms. a A serrated lesion characterized by serrated crypt profiles. b Transition phase from a serrated lesion with dysplasia (left) to a low-grade mucinous neoplasm (right). c, d Different phenotypic appearance of LAMN. e HAMN showing a complex pattern of growth. f p53 immunohistochemical expression of the neoplastic glands of the lesion showed in e. Original magnifications ×10, ×20 and ×40
Neoplastic areas were manually microdissected from 10 μm unstained histological sections formalin-fixed and paraffin-embedded (FFPE) samples using the original haematoxylin and eosin-stained slide as guide to select the region of interest. The DNA was extracted from each target using the QIAamp DNA FFPE tissue kit (Qiagen, Milan, Italy) according to the manufacturer’s instructions.
RNF43 and SMAD4 mutational analysis
The DNA isolated from tumour samples was used to perform PCR amplification of RNF43 (exons 2, 3, 4, 5, 6, 7 and 8) and SMAD4 (exons 1, 2 and 3) genes. Amplified PCR products were purified and sequenced by the Sanger method. Primers used for PCR and Sanger sequencing are available upon request.
Hotspot KRAS, NRAS, BRAF and PIK3CA genes mutational profiling
The mutational status of KRAS, NRAS, BRAF and PIK3CA hotspot regions was assessed using the high-throughput genotyping platform Sequenom MassARRAY System (Agena Biosciences, San Diego, CA, USA) and the Myriapod Colon Status Kit (Diatech Pharmacogenetics, Jesi, Italy), following the manufacturer’s instructions. This molecular array allows to identify the most relevant mutations of KRAS (codons 12, 13, 59, 61, 117, and 146), NRAS (codons 12, 13, 18, 59, 61, 117, and 146), BRAF (codons 594, 600, and 601), and PIK3CA genes (codons 38, 81, 88, 93, 108, 118, 345, 420, 539, 542, 545, 546, 549, 1021, 1025, 1043, 1047, and 1049).
Immunohistochemical analysis
Immunohistochemical staining for p53 (clone DO-7; Agilent, Santa Clara, CA; prediluited), MLH1 (clone ES05; Agilen; dilution 1:25), PMS2 (clone EP51; Agilent; dilution 1:20); MSH2 (clone FE11; Agilent; dilution 1:50), and MSH6 (clone EP49; Agilent; dilution 1:25) was performed on a Leica Bond system (Bond-III; Leica Microsystems, Buccinasco, Milan, Italy). The slides were jointly assessed by two pathologists. Nuclear immunostaining for MLH1, PMS2, MSH2, and MSH6 was evaluated following the AIFEG-SIAPeC criteria to identify mismatch repair-deficiency (MMRd) and mismatch repair-proficiency (MMRp) (Remo et al. 2016). Expression of p53 was considered as “altered” in the presence of more than 30% neoplastic nuclei showing a strong staining or in the case of complete absence in the context of a positive background.
Results
Clinico-pathological features of the series
Samples obtained from 49 Caucasian patients were included in the study. Specifically, appendectomy was performed for PMP in 2 cases, during CRC surgery in 32 cases, abdominal surgery for extra-colic neoplastic diseases in 5 cases (1 IPMN, 1 uterine adenocarcinoma, 1 small bowel adenocarcinoma and 2 gallbladder cancer). For 3 cases, patients underwent appendectomy secondary to bowel surgery for non-neoplastic disease (i.e. ileal resection due to umbilical hernia, treatment of not-responsive ulcerative colitis, anastomotic dehiscence). Seven patients required appendiceal removal because of appendiceal pathology (1 mucocele, 1 urgent intervention for intestinal obstruction and 5 acute appendicitis).
For three cases, two separate lesions were microdissected from the same appendix: the first case had the concomitant presence of a hyperplastic polyp (HP) and a tubulo-villous adenoma; in the second case, the coexisting LAMN and HAMN features were microdissected and analysed separately; in the third case, the mucinous adenocarcinoma with its adjacent high-grade tubular adenoma were microdissected separately.
Molecular landscape of appendicular lesions (Fig. 2)
a Graphic representation of the results of the molecular profiling of mucinous appendicular neoplasia. On the top, horizontally, the type of lesion is reported (colors legend on the right). Vertically, on the left, the names of mutated/altered genes (italics, capital letters) or proteins with altered expression is shown. Alterations are indicated by red squares. The prevalence of molecular alterations is displayed on the right. HP hyperplastic polyp, TVLG tubulo-villous adenoma with low-grade dysplasia, THG tubular adenoma with high-grade dysplasia, SSL sessile serrated lesion, SSLd sessile serrated lesion with dysplasia, LAMN low grade appendiceal mucinous neoplasm, HAMN high grade appendiceal mucinous neoplasm, ADK mucinous adenocarcinoma, MMRd DNA mismatch repair deficiency. b Representative examples of Sanger sequencing for a mutation identified in the RNF43 gene. Left electropherogram with the representation of the p.R117H mutated forward sequence of the RNF43 gene and its respective reverse sequence (bottom right)
Matched lesions
Of the three matched samples, the patient carrying concomitant HP and tubulo-villous adenoma with low-grade dysplasia; showed absence of mutations in the genes analysed, normal p53 expression and non-altered microsatellite status. In the second patient, affected by simultaneous LAMN and HAMN, both samples revealed a common driver KRAS p.G12D point mutation and the altered p53 expression. Only data on the HAMN lesion were retained for mutational prevalence and Fig. 2 output. The third case included a mucinous adenocarcinoma and the tubular adenoma from which the carcinoma initiated; both lesions resulted wild type for the tested genes, except for a p.G12V point mutation in the KRAS gene.
Appendiceal mucinous adenocarcinomas
Our results showed that KRAS was the main mutated gene in appendiceal mucinous adenocarcinoma, especially in exon 2 (4/9 cases), followed by RNF43 (3/9). One case of appendiceal mucinous adenocarcinoma had BRAF p.V600E mutation and alteration in the RNF43 gene with stable expression of MMR proteins. This was the only case showing strong IHC positivity for p53. NRAS gene was mutated in one case.
High-grade appendiceal mucinous neoplasms (HAMN)
Two (66.67%) out of 3 cases had codon 12 of exon 2 KRAS gene mutation, and one of them had p53 dysregulation.
Two out of three cases had alterations in the RNF43 gene. The first had an alteration in RNF43 p.R296C in concomitance with the mutations in KRAS and NRAS; the second case showed the RNF43 p.R117H mutation and p53 dysregulation with wild type BRAF/RAS genes.
Low-grade appendiceal mucinous neoplasms (LAMN)
The majority of LAMNs showed alterations in KRAS: 55.56% (10/18) in exon 2 and 5.55% (1/18) in exon 4. One case had a mutation in BRAF p.V600E (5.6%). Another case showed the concurrent presence of mutations in KRAS and BRAF. In addition, 4/18 LAMN were wild type for the entire gene panel. An altered p53 IHC expression was observed in five lesions.
Serrated sessile lesions (SSL) and serrated lesions with dysplasia (SSLd)
8/15 (53.33%) SSL harboured KRAS main gene mutations: 6/15 cases (40%) in exon 2 and 2/15 cases (13.33%) in the exons 3 and 4. Furthermore, 4/15 cases (26.67%) harboured BRAF p.V600E mutation. Three cases were SSLd; 2/3 (66.67%) had exon 2 KRAS mutation; 1/3 (33.33%) was BRAF/RAS wild type. One case showed the simultaneous presence of mutations in KRAS and RNF43.
An altered IHC p53 expression was observed in three SSL lesions. One SSLd also showed strong IHC positivity for p53.
One case resulted mutated in exon 5 of the RNF43 gene. One case of SSL was RNF43 p.C222Y mutated and alteration in p53 expression was detected by IHC.
Other lesions
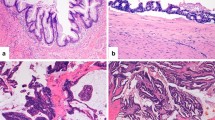
The tubular adenoma with high-grade dysplasia was characterized by a BRAF p.V600E mutation and mismatch repair deficiency (loss of MLH1/PMS2) (Fig. 3). Of note, this was the only case of microsatellite instability found in the entire series analysed. The tubulo-villous adenoma resulted wild type for all genes investigated. The only HP was wild type for all the tested genes, MMRp and with retained p53 expression.
A tubular adenoma with high-grade dysplasia showing a peculiar molecular profile for appendicular mucinous neoplasm. a Gross characteristics of the lesion. b High grade dysplastic glands. c, d Representative Sequenom MassArray output profile and Sanger chromatogram of the p.V600E BRAF mutation. e Loss of PMS2 in the epithelial neoplastic cells. Of note, this was the only case with MMRd of the entire series
Discussion
This study molecularly profiled a relatively large spectrum of preinvasive and mucinous neoplasms of the appendix.
Consistently with the literature, KRAS mutations predominated among the different appendiceal lesions: in fact, half of our cases showed KRAS point mutations and these mutations harboured in SSLs (about 60%) as well as in adenocarcinomas (44%). KRAS mutations (preferentially observed in exon 2) seem to have a pivotal role in the development of appendiceal mucinous neoplasms (Kabbani et al. 2002; Borazanci et al. 2017). KRAS is mutated in 41–100% of appendiceal mucinous adenomas (Szych et al. 1999; Zauber et al. 2011; Yantiss et al. 2007; Tsai et al. 2019; Liao et al. 2020; Yanai et al. 2020). Pai et al. (2014) analysed a series of 132 appendiceal lesions, revealing that serrated lesions of the appendix often harbour KRAS mutations and only infrequently display BRAF mutations. Moreover, KRAS mutations have been identified in a high proportion of disseminated mucinous neoplasms (Tokunaga et al. 2019; Davison et al. 2014; Szych et al. 1999; Zauber et al. 2011; Tsai et al. 2019). Notably, Davison and colleagues (Davison et al. 2014) have reported that, in their series, none of the high-grade mucinous adenocarcinomas with a predominant signet ring cell component had KRAS mutation.
All these findings support the idea that KRAS may be biologically more important than BRAF in appendiceal lesions, and the serrated pathway of carcinogenesis may have less relevance in the appendix than in the colon (WHO Classification of Tumours Editorial Board 2019; Carr et al. 2009; Murakami et al. 2018). We have detected BRAF mutations in 13% of our cases. Our findings are quite different from previous data reported in the literature, which pinpointed a relative rarity of BRAF mutations in appendicular SSLs (Pai et al. 2014), with the exception of Tsai and colleagues (Tsai et al. 2019) that found 78%(7/9) of serrated polyps characterized by a BRAF p.V600E mutation. KRAS and BRAF mutations are mutually exclusive in CRC: the shortage of BRAF mutations detected in appendiceal neoplasms with PMP seems to be a consistent consequent high prevalence of KRAS mutations among appendiceal mucinous neoplasms (Tokunaga et al. 2019).
Another frequently mutated gene in appendiceal tumours is GNAS (Ang et al. 2018; Alakus et al. 2014), which was not analysed in our series. Curiously, GNAS mutations coexist with the KRAS mutation in 65–85% of cases (Gleeson et al. 2018; Stein et al. 2020). Liu and colleagues (Liu et al. 2014) reported KRAS and GNAS are the most commonly mutated genes in LAMNs and low-grade mucinous adenocarcinomas with PMP. According to Ang and colleagues (Ang et al. 2018), GNAS mutations are usually absent in high-grade neoplasms, while mutations in TP53 are usually absent in low-grade ones. These findings outline a significant association of GNAS mutation with low-grade tumours and of TP53 mutation with high-grade tumours, suggesting the majority of high-grade appendiceal tumours occur de novo, rather than progressing from low-grade neoplasms (Ang et al. 2018; Alakus et al. 2014). Conversely, Hara and colleagues (Hara et al. 2015) reported that LAMNs and appendiceal mucinous carcinomas might share a mutational spectrum comprising KRAS, TP53 and GNAS genes, suggesting that mucinous carcinomas might evolve from LAMNs. However, Singhi and colleagues (Singhi et al. 2014) showed that GNAS is commonly mutated in both low-grade and high-grade disseminated appendiceal mucinous neoplasms, suggesting that GNAS mutation status is not related to the grade of appendiceal mucinous neoplasms. Similarly, Liao and colleagues (Liao et al. 2020) recently showed that HAMN and LAMN share high rates of KRAS and GNAS co-mutations supporting a common histogenesis and distinguishing them from mucinous adenocarcinoma, which is characterized by KRAS mutations in the absence of GNAS alterations.
RNF43 mutations were present in 7 cases (13%), particularly in 3 adenocarcinomas, 2 LAMNs and 2 HAMN. These data support an important role of this mutation in appendiceal neoplasms progression. This seem to be in agreement with previously published data, since alterations of RNF43 affect both up-stream (Koo et al. 2012) and down-stream (Loregger et al. 2015) activation of Frizzled and consequently of Wnt/beta-catenin pathway (Borowsky et al. 2018; Hao et al. 2016). Yanai and colleagues (Yanai et al. 2020) recently suggested RNF43 mutations may occur at a later stage of mucinous adenocarcinoma development and may not be associated with PMP. The significance of RNF43 mutation in appendiceal tumours has not been clarified yet, but its role has been better characterized in IPMN (Lee et al. 2016). This is not the only mutation that connects pancreatic and appendicular neoplasia: indeed, KRAS and GNAS mutations have been found in both lesions, suggesting similarities in these two cancerogenic pathways (Lee et al. 2016; Reid et al. 2014; Fischer and Wood 2018; Sakamoto et al. 2015). Similarly, this could explain why abnormal p53 expression by immunohistochemistry is reported in only 21% of our cases. Moreover, prominent mucin production is a common phenotypic feature of LAMN and IPMN (Nishikawa et al. 2013).
Several studies (Valasek and Pai 2018; Nummela et al. 2015) have identified abnormal TP53 expression in a significant fraction (30%) of high-grade mucinous adenocarcinoma. Considering that only 7% of low-grade tumours show this characteristic, TP53 status appears to be the only marker associated with the acquisition of an aggressive phenotype in tumours with PMP. We reported an altered immunohistochemical expression of p53 in 13.5% of our cases.
The role of DNA mismatch repair (MMR) in appendix mucosa tumours remains elusive. To the best of our knowledge, only a single study showed a significant prevalence of microsatellite instability in appendicular neoplastic pathology, but most of the tumours tested were characterized by a non-mucinous histotype (Taggart et al. 2013). Moreover, Tokunaga and colleagues (Tokunaga et al. 2019) suggested the immune profile of appendiceal adenocarcinomas is similar to left-sided CRCs but not to right-sided CRCs. Coherently with the literature (Misdraji et al. 2004), genes that code for proteins responsible for the maintenance of microsatellite stability (i.e. expression of the MMR proteins) are not affected in appendicular carcinogenesis: MMR proteins are fully expressed in mucinous neoplasm of the appendix, independently of the histological subtype. This fact suggests that microsatellite instability has no (or minimal) etiological role in the development of appendicular mucinous tumours. In our study, only one case showed MMRd and it was characterized by a V600E BRAF mutation, as observed in sporadic MMRd CRCs.
In conclusion, despite the similar phenotype, many non-invasive appendicular mucinous lesions are characterized by a different genetic landscape in comparison to colorectal serrated lesions. The DNA mismatch repair complex seems not to be altered within appendiceal mucinous carcinogenesis.
Availability of data and material
Upon request.
References
Alakus H, Babicky ML, Ghosh P, Yost S, Jepsen K, Dai Y et al (2014) Genome-wide mutational landscape of mucinous carcinomatosis peritonei of appendiceal origin. Genome Med 6(5):43. https://doi.org/10.1186/gm559
Ang CS, Shen JP, Hardy-Abeloos CJ, Huang JK, Ross JS, Miller VA et al (2018) Genomic landscape of appendiceal neoplasms. JCO Precis Oncol. https://doi.org/10.1200/PO.17.00302
Borazanci E, Millis SZ, Kimbrough J, Doll N, Von Hoff D, Ramanathan RK (2017) Potential actionable targets in appendiceal cancer detected by immunohistochemistry, fluorescent in situ hybridization, and mutational analysis. J Gastrointest Oncol 8(1):164–172. https://doi.org/10.21037/jgo.2017.01.14
Borowsky J, Dumenil T, Bettington M, Pearson SA, Bond C, Fennell L et al (2018) The role of APC in WNT pathway activation in serrated neoplasia. Mod Pathol 31(3):495–504. https://doi.org/10.1038/modpathol.2017.150
Carr NJ, Mahajan H, Tan KL, Hawkins NJ, Ward RL (2009) Serrated and non-serrated polyps of the colorectum: their prevalence in an unselected case series and correlation of BRAF mutation analysis with the diagnosis of sessile serrated adenoma. J Clin Pathol 62(6):516–518. https://doi.org/10.1136/jcp.2008.061960
Carr NJ, Finch J, Ilesley IC, Chandrakumaran K, Mohamed F, Mirnezami A et al (2012) Pathology and prognosis in pseudomyxoma peritonei: a review of 274 cases. J Clin Pathol 65(10):919–923. https://doi.org/10.1136/jclinpath-2012-200843
Carr NJ, Cecil TD, Mohamed F, Sobin LH, Sugarbaker PH, González-Moreno S et al (2016) A consensus for classification and pathologic reporting of pseudomyxoma peritonei and associated appendiceal neoplasia: the results of the Peritoneal Surface Oncology Group International (PSOGI) Modified Delphi Process. Am J Surg Pathol 40(1):14–26. https://doi.org/10.1097/PAS.0000000000000535
Connor SJ, Hanna GB, Frizelle FA (1998) Appendiceal tumors: retrospective clinicopathologic analysis of appendiceal tumors from 7,970 appendectomies. Dis Colon Rectum 41(1):75–80. https://doi.org/10.1007/BF02236899
Davison JM, Choudry HA, Pingpank JF, Ahrendt SA, Holtzman MP, Zureikat AH et al (2014) Clinicopathologic and molecular analysis of disseminated appendiceal mucinous neoplasms: identification of factors predicting survival and proposed criteria for a three-tiered assessment of tumor grade. Mod Pathol 27(11):1521–1539. https://doi.org/10.1038/modpathol.2014.37
Fischer CG, Wood LD (2018) From somatic mutation to early detection: insights from molecular characterization of pancreatic cancer precursor lesions. J Pathol 246(4):395–404. https://doi.org/10.1002/path.5154
Gleeson EM, Feldman R, Mapow BL, Mackovick LT, Ward KM, Morano WF et al (2018) Appendix-derived pseudomyxoma peritonei (PMP): molecular profiling toward treatment of a rare malignancy. Am J Clin Oncol 41(8):777–783. https://doi.org/10.1097/COC.0000000000000376
Hao HX, Jiang X, Cong F (2016) Control of Wnt receptor turnover by R-spondin-ZNRF3/RNF43 signaling module and its dysregulation in cancer. Cancers (Basel). https://doi.org/10.3390/cancers8060054
Hara K, Saito T, Hayashi T, Yimit A, Takahashi M, Mitani K et al (2015) A mutation spectrum that includes GNAS, KRAS and TP53 may be shared by mucinous neoplasms of the appendix. Pathol Res Pract 211(9):657–664. https://doi.org/10.1016/j.prp.2015.06.004
Kabbani W, Houlihan PS, Luthra R, Hamilton SR, Rashid A (2002) Mucinous and nonmucinous appendiceal adenocarcinomas: different clinicopathological features but similar genetic alterations. Mod Pathol 15(6):599–605. https://doi.org/10.1038/modpathol.3880572
Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M et al (2012) Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488(7413):665–669. https://doi.org/10.1038/nature11308
Lee JH, Kim Y, Choi JW, Kim YS (2016) KRAS, GNAS, and RNF43 mutations in intraductal papillary mucinous neoplasm of the pancreas: a meta-analysis. Springerplus 5(1):1172. https://doi.org/10.1186/s40064-016-2847-4
Liao X, Vavinskaya V, Sun K, Hao Y, Li X, Valasek M et al (2020) Mutation profile of high-grade appendiceal mucinous neoplasm. Histopathology 76(3):461–469. https://doi.org/10.1111/his.13986
Liu X, Mody K, de Abreu FB, Pipas JM, Peterson JD, Gallagher TL et al (2014) Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin Chem 60(7):1004–1011. https://doi.org/10.1373/clinchem.2014.225565
Loregger A, Grandl M, Mejías-Luque R, Allgäuer M, Degenhart K, Haselmann V et al (2015) The E3 ligase RNF43 inhibits Wnt signaling downstream of mutated β-catenin by sequestering TCF4 to the nuclear membrane. Sci Signal 8(393):ra90. https://doi.org/10.1126/scisignal.aac6757
Misdraji J, Burgart LJ, Lauwers GY (2004) Defective mismatch repair in the pathogenesis of low-grade appendiceal mucinous neoplasms and adenocarcinomas. Mod Pathol 17(12):1447–1454. https://doi.org/10.1038/modpathol.3800212
Murakami T, Akazawa Y, Yatagai N, Hiromoto T, Sasahara N, Saito T et al (2018) Molecular characterization of sessile serrated adenoma/polyps with dysplasia/carcinoma based on immunohistochemistry, next-generation sequencing, and microsatellite instability testing: a case series study. Diagn Pathol 13(1):88. https://doi.org/10.1186/s13000-018-0771-3
Nishikawa G, Sekine S, Ogawa R, Matsubara A, Mori T, Taniguchi H et al (2013) Frequent GNAS mutations in low-grade appendiceal mucinous neoplasms. Br J Cancer 108(4):951–958. https://doi.org/10.1038/bjc.2013.47
Nummela P, Saarinen L, Thiel A, Järvinen P, Lehtonen R, Lepistö A et al (2015) Genomic profile of pseudomyxoma peritonei analyzed using next-generation sequencing and immunohistochemistry. Int J Cancer 136(5):E282–E289. https://doi.org/10.1002/ijc.29245
Pai RK, Hartman DJ, Gonzalo DH, Lai KK, Downs-Kelly E, Goldblum JR et al (2014) Serrated lesions of the appendix frequently harbor KRAS mutations and not BRAF mutations indicating a distinctly different serrated neoplastic pathway in the appendix. Hum Pathol 45(2):227–235. https://doi.org/10.1016/j.humpath.2013.10.021
Pietrantonio F, Maggi C, Fanetti G, Iacovelli R, Di Bartolomeo M, Ricchini F et al (2014) FOLFOX-4 chemotherapy for patients with unresectable or relapsed peritoneal pseudomyxoma. Oncologist 19(8):845–850. https://doi.org/10.1634/theoncologist.2014-0106
Reghunathan M, Kelly KJ, Valasek MA, Lowy AM, Baumgartner JM (2018) Histologic predictors of recurrence in mucinous appendiceal tumors with peritoneal dissemination after HIPEC. Ann Surg Oncol 25(3):702–708. https://doi.org/10.1245/s10434-017-6310-8
Reid MD, Saka B, Balci S, Goldblum AS, Adsay NV (2014) Molecular genetics of pancreatic neoplasms and their morphologic correlates: an update on recent advances and potential diagnostic applications. Am J Clin Pathol 141(2):168–180. https://doi.org/10.1309/AJCP0FKDP7ENVKEV
Remo A, Fassan M, Lanza G (2016) Immunohistochemical evaluation of mismatch repair proteins in colorectal carcinoma: the AIFEG/GIPAD proposal. Pathologica 108(3):104–109
Sakamoto H, Kuboki Y, Hatori T, Yamamoto M, Sugiyama M, Shibata N et al (2015) Clinicopathological significance of somatic RNF43 mutation and aberrant expression of ring finger protein 43 in intraductal papillary mucinous neoplasms of the pancreas. Mod Pathol 28(2):261–267. https://doi.org/10.1038/modpathol.2014.98
Singhi AD, Davison JM, Choudry HA, Pingpank JF, Ahrendt SA, Holtzman MP et al (2014) GNAS is frequently mutated in both low-grade and high-grade disseminated appendiceal mucinous neoplasms but does not affect survival. Hum Pathol 45(8):1737–1743. https://doi.org/10.1016/j.humpath.2014.04.018
Smeenk RM, van Velthuysen ML, Verwaal VJ, Zoetmulder FA (2008) Appendiceal neoplasms and pseudomyxoma peritonei: a population based study. Eur J Surg Oncol 34(2):196–201. https://doi.org/10.1016/j.ejso.2007.04.002
Stein A, Strong E, Clark Gamblin T, Clarke C, Tsai S, Thomas J et al (2020) Molecular and genetic markers in appendiceal mucinous tumors: a systematic review. Ann Surg Oncol 27(1):85–97. https://doi.org/10.1245/s10434-019-07879-7
Szych C, Staebler A, Connolly DC, Wu R, Cho KR, Ronnett BM (1999) Molecular genetic evidence supporting the clonality and appendiceal origin of Pseudomyxoma peritonei in women. Am J Pathol 154(6):1849–1855. https://doi.org/10.1016/S0002-9440(10)65442-9
Taggart MW, Galbincea J, Mansfield PF, Fournier KF, Royal RE, Overman MJ et al (2013) High-level microsatellite instability in appendiceal carcinomas. Am J Surg Pathol 37(8):1192–1200. https://doi.org/10.1097/PAS.0b013e318282649b
Tokunaga R, Xiu J, Johnston C, Goldberg RM, Philip PA, Seeber A et al (2019) Molecular profiling of appendiceal adenocarcinoma and comparison with right-sided and left-sided colorectal cancer. Clin Cancer Res 25(10):3096–3103. https://doi.org/10.1158/1078-0432.CCR-18-3388
Tsai JH, Yang CY, Yuan RH, Jeng YM (2019) Correlation of molecular and morphological features of appendiceal epithelial neoplasms. Histopathology 75(4):468–477. https://doi.org/10.1111/his.13924
Valasek MA, Pai RK (2018) An update on the diagnosis, grading, and staging of appendiceal mucinous neoplasms. Adv Anat Pathol 25(1):38–60. https://doi.org/10.1097/PAP.0000000000000178
WHO Classification of Tumours Editorial Board (2019) Digestive system tumours. International Agency for Research on Cancer, Lyon
Yanai Y, Saito T, Hayashi T, Akazawa Y, Yatagai N, Tsuyama S et al (2020) Molecular and clinicopathological features of appendiceal mucinous neoplasms. Virchows Arch. https://doi.org/10.1007/s00428-020-02906-5
Yantiss RK, Panczykowski A, Misdraji J, Hahn HP, Odze RD, Rennert H et al (2007) A comprehensive study of nondysplastic and dysplastic serrated polyps of the vermiform appendix. Am J Surg Pathol 31(11):1742–1753. https://doi.org/10.1097/PAS.0b013e31806bee6d
Zauber P, Berman E, Marotta S, Sabbath-Solitare M, Bishop T (2011) Ki-ras gene mutations are invariably present in low-grade mucinous tumors of the vermiform appendix. Scand J Gastroenterol 46(7–8):869–874. https://doi.org/10.3109/00365521.2011.565070
Funding
Open access funding provided by Università degli Studi di Padova within the CRUI-CARE Agreement. This work was partly supported by a Grant from the Italian Health Ministry’s research program NET-2016–02363853. The funding agency had no role in the design and performance of the study.
Author information
Authors and Affiliations
Contributions
GM, GB, PM, CB, FG, EB, CL conceived and carried out the experiments. GP, MS, SP, GS, CM, AS, SL, FL contributed to sample preparation. FL, CL, APDT, MF contributed to the interpretation of the results. GM, GB, PM, MF took the lead in writing the manuscript. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare related to the presented work.
Ethics approval
All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Munari, G., Businello, G., Mattiolo, P. et al. Molecular profiling of appendiceal serrated lesions, polyps and mucinous neoplasms: a single-centre experience. J Cancer Res Clin Oncol 147, 1897–1904 (2021). https://doi.org/10.1007/s00432-021-03589-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-021-03589-4