Abstract
Background
This study was designed to detect patients with early NSCLC with tentatively using the stem signatures associated autoantibodies (AAbs), and to evaluate its latent values in the early diagnosis and precise prognosis prediction.
Methods
The serum concentrations of selective antibodies were quantitated by enzyme-linked immunosorbent assay (ELISA), and a total of 458 cases were enrolled (training set = 401; validation set = 57). TCGA databases were used to analyze the distinct expressions and prognostic values of related genes. The optimal cut-off values were 11.60 U/ml for P53, 4.90 U/ml for MAGEA1, 3.85 U/ml for SOX2, and 7.05U/ml for PGP9.5.
Results
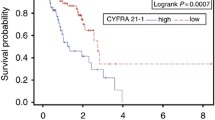
We found that the stem signatures associated antibodies of MAGEA1, PGP9.5, SOX2, and TP53 exhibited high expressions in NSCLC, negatively correlating with the overall survival (OS) (P < 0.05). In the test groups, the diagnosis sensitivity of P53, PGP9.5, SOX2, and MAGEA1 reached to 21.5%, 39.0%, 50.3%, and 35.0%, respectively, and the specificity reached to 98.7%, 99.4%, 92.2%, and 97.4%. The four candidates’ panel gave a sensitivity of 71.8% with a specificity of 89%. In the validation group, the detection of the four antibodies in early diagnosis of NSCLC also exhibited high specificity and sensitivity, further consolidating their potential application.
Conclusions
The detection regarding stem signatures associated antibodies could be used as effective tools in early NSCLC diagnosis, but not for localized screening of cancers, and their abnormal expression was in accordance with poorer survival.





Similar content being viewed by others
References
Bach PB et al (2007) Computed tomography screening and lung cancer outcomes. JAMA 297(9):953–961
Bach PB et al (2012) Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 307(22):2418–2429
Blandin Knight S et al (2017) Progress and prospects of early detection in lung cancer. Open Biol 7(9)
Boyle P et al (2011) Clinical validation of an autoantibody test for lung cancer. Ann Oncol 22(2):383–389
Chapman CJ et al (2008) Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax 63(3):228–233
Chapman CJ et al (2012) EarlyCDT(R)-Lung test: improved clinical utility through additional autoantibody assays. Tumour Biol 33(5):1319–1326
Croswell JM et al (2010) (2010) Cumulative incidence of false-positive test results in lung cancer screening: a randomized trial. Ann Intern Med 152(8):505–512 (W176-80)
Grah JJ et al (2014) Clinical significance of immunohistochemical expression of cancer/testis tumor-associated antigens (MAGE-A1, MAGE-A3/4, NY-ESO-1) in patients with non-small cell lung cancer. Tumori 100(1):60–68
Gyorffy B et al (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 8(12):e82241
Huang G et al (2019) TUSC7 suppression of Notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cells. Life Sci 232:116630
Kossenkov AV et al (2019) A gene expression classifier from whole blood distinguishes benign from malignant lung nodules detected by low-dose CT. Cancer Res 79(1):263–273
Kulpa J et al (2002) Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21–1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem 48(11):1931–1937
Lam S et al (2011) EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 4(7):1126–1134
Li P et al (2017) Evaluation of serum autoantibodies against tumor-associated antigens as biomarkers in lung cancer. Tumour Biol 39(10):1010428317711662
Mauro C et al (2019) New and old biomarkers in the differential diagnosis of lung cancer: pro-gastrin-releasing peptide in comparison with neuron-specific enolase, carcinoembryonic antigen, and CYFRA 21-1. Int J Biol Mark 34(2):163–167. https://doi.org/10.1177/1724600819834235
Mazzone PJ et al (2018) Evaluation of a serum lung cancer biomarker panel. Biomark Insights 13:1177271917751608
Murray A et al (2010) Technical validation of an autoantibody test for lung cancer. Ann Oncol 21(8):1687–1693
Nakamura H, Nishimura T (2017) History, molecular features, and clinical importance of conventional serum biomarkers in lung cancer. Surg Today 47(9):1037–1059
National Lung Screening Trial Research T et al (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365(5):395–409
Oudkerk M et al (2017) European position statement on lung cancer screening. Lancet Oncol 18(12):e754–e766
Rhodes DR et al (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6(1):1–6
Rhodes DR et al (2007) Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9(2):166–180
Roberts H et al (2013) Screening high-risk populations for lung cancer: guideline recommendations. J Thorac Oncol 8(10):1232–1237
Seijo LM et al (2019) Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol 14(3):343–357
Siegel RL, Miller KD, Jemal A (2017) Cancer Statistics, 2017. CA Cancer J Clin 67(1):7–30
Sun X et al (2015) MiR-208a stimulates the cocktail of SOX2 and beta-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget 6(32):32944–32954
Tang Z et al (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102
Vargas AJ, Harris CC (2016) Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer 16(8):525–537
Vatankulu B et al (2016) Accuracy of FDG-PET/CT and paraneoplastic antibodies in diagnosing cancer in paraneoplastic neurological syndromes. Rev Esp Med Nucl Imagen Mol 35(1):17–21
Wang M et al (2019) H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif 52(1):e12534
Wang H et al (2020) Long noncoding RNA LINC01116 contributes to gefitinib resistance in non-small cell lung cancer through regulating IFI44. Mol Ther Nucl Acids 19:218–227
Xiao G et al (2019) FAM83A-AS1 promotes lung adenocarcinoma cell migration and invasion by targeting miR-150-5p and modifying MMP14. Cell Cycle 18(21):2972–2985
Yao Y et al (2012) Potential application of non-small cell lung cancer-associated autoantibodies to early cancer diagnosis. Biochem Biophys Res Commun 423(3):613–619
Acknowledgements
The authors acknowledge assistants in the Center for Translational Medicine of First Affiliated Hospital of Xi’an Jiaotong University, for their technical assistance. The authors are very appreciative of the great help they received from the staffs of the Thoracic Department and Oncology Department. This experiment was supported by the National Science Foundation for Young Scientists of China, grant No. 81602597 (Referred to Xin Sun), and Foundation Research Project of Shaanxi Province (The Natural Science Fund No. 2018JM7017 (Referred to Xin Sun).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflicts of interest in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
432_2020_3325_MOESM1_ESM.tif
Figure S1 Expression patterns of Stem signatures associated antibodiesThe stem cells potency associated UCHL1, SOX2, TP53, MAGEA1, SOX4, WNT1, TCF4, NOTCH1, KLF4, MYC, LIN28A, LIN28B, SNAI1, PROM1, ALDH1A1, CD44, CREBBP, were all applied for expression, and the results were exhibited in heatmap. Detections regarding to stem cells potency associated members were universally deregulated, enhanced with mutated or overexpressed status
432_2020_3325_MOESM2_ESM.tif
Figure S2 Protein expression patterns of Stem signatures associated antibodies A. Strong cytoplasmic staining of PGP9.5 was found in cases of gliomas, malignant, testis, cervical and lung cancers. The positivity was often accompanied with weak to moderate nuclear immunoreactivity. B. Strong nuclear positivity of SOX2 was mainly observed in several cases of glioma, testis cancer and squamous cell carcinoma. C. Many malignant cells displayed moderate to strong nuclear positivity of TP53. D. Moderate to string cytoplasmic and nuclear immunoreactivity of MAGEA1 was observed in a few cases of testicular, skin, lung and head & neck cancers. Remaining malignant cells were generally negative
432_2020_3325_MOESM3_ESM.tif
Figure S3 The expressions of four Stem signatures associated genes in multiple malignanciesThe graph shows the number of analyses meeting the threshold with statistically significant over-expression (red) or down-regulated expression (blue) of the target gene. Cell color is determined by the best gene rank percentile for the analyses within the cell. (A) MAGEA1, (B) PGP9.5, (C) SOX2, (D) TP53 expressions associated genes in multiple malignancies
432_2020_3325_MOESM4_ESM.tif
Figure S4 Expressions of four genes in LUAD and LUSC Box plots were used to compare the distinct expressions of MAGEA1 (A), PGP9.5 (B), SOX2 (C) and TP53 (D), either between LUSC and normal tissues, or between LUAD and normal tissues, by analyzing the GEPIA database. The threshold was set as Log2FC=1, p = 0.05. T was tumor. N was normal. * P value <0.05.
Rights and permissions
About this article
Cite this article
Chen, SS., Li, K., Wu, J. et al. Stem signatures associated antibodies yield early diagnosis and precise prognosis predication of patients with non-small cell lung cancer. J Cancer Res Clin Oncol 147, 223–233 (2021). https://doi.org/10.1007/s00432-020-03325-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-020-03325-4