Abstract
Purpose
Uterine neoplasms comprise a broad spectrum of lesions, some of which may pose a diagnostic challenge even to experienced pathologists. Recently, genome-wide DNA methylation-based classification of central nervous system tumors has been shown to increase diagnostic precision in clinical practice when combined with standard histopathology. In this study, we describe DNA methylation patterns of a diverse set of uterine neoplasms and test the applicability of array-based DNA methylation profiling.
Methods
A multicenter cohort including prototypical epithelial and mesenchymal uterine neoplasms was collected. Tumors were subject to pathology review and array-based DNA methylation profiling (Illumina Infinium HumanMethylation450 or EPIC [850k] BeadChip). Methylation data were analyzed by unsupervised hierarchical clustering and t-SNE analysis.
Results
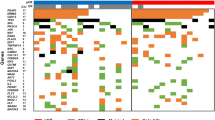
After sample retrieval and pathology review the study cohort consisted of 49 endometrial carcinomas (EC), 5 carcinosarcomas (MMMT), 8 uterine leiomyomas (ULMO), 7 uterine leiomyosarcomas (ULMS), 15 uterine tumor resembling ovarian sex cord tumors (UTROSCT), 17 low-grade endometrial stromal sarcomas (LGESS) and 9 high-grade endometrial stromal sarcomas (HGESS). Analysis of methylation data identified distinct methylation clusters, which correlated with established diagnostic categories of uterine neoplasms. MMMT clustered together with EC, while ULMO, ULMS and UTROSCT each formed distinct clusters. The LGESS cluster differed from that of HGESS, and within the branch of HGESS, we observed a notable subgrouping of YWHAE- and BCOR-rearranged tumors.
Conclusion
Herein, we describe distinct DNA methylation signatures in uterine neoplasms and show that array-based DNA methylation analysis holds promise as an ancillary tool to further characterize uterine neoplasms, especially in cases which are diagnostically challenging by conventional techniques.


Similar content being viewed by others
References
Altrabulsi B, Malpica A, Deavers MT, Bodurka DC, Broaddus R, Silva EG (2005) Undifferentiated carcinoma of the endometrium. Am J Surg Pathol 29(10):1316–1321
Bodelon C, Killian JK, Sampson JN, Anderson WF, Matsuno R, Brinton LA, Wentzensen N (2019) Molecular classification of epithelial ovarian cancer based on methylation profiling: evidence for survival heterogeneity. Clin Cancer Res. https://doi.org/10.1158/1078-0432.CCR-18-3720
Brooks RA, Fleming GF, Lastra RR, Lee NK, Moroney JW, Son CH, Veneris JL (2019) Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin 69(4):258–279. https://doi.org/10.3322/caac.21561
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Pfister SM (2018) DNA methylation-based classification of central nervous system tumours. Nature 555(7697):469–474. https://doi.org/10.1038/nature26000
Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, Levine DA (2017) integrated molecular characterization of uterine carcinosarcoma. Cancer Cell 31(3):411–423. https://doi.org/10.1016/j.ccell.2017.02.010
Chiang S, Lee CH, Stewart CJR, Oliva E, Hoang LN, Ali RH, Soslow RA (2017) BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod Pathol 30(9):1251–1261. https://doi.org/10.1038/modpathol.2017.42
Dickson BC, Childs TJ, Colgan TJ, Sung YS, Swanson D, Zhang L, Antonescu CR (2019) Uterine tumor resembling ovarian sex cord tumor: a distinct entity characterized by recurrent NCOA2/3 gene fusions. Am J Surg Pathol 43(2):178–186. https://doi.org/10.1097/PAS.0000000000001153
Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8(4):286–298. https://doi.org/10.1038/nrg2005
Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan G, Wang CY (2009) BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol 11(8):1002–1009. https://doi.org/10.1038/ncb1913
Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Esteller M (2012) A DNA methylation fingerprint of 1628 human samples. Genome Res 22(2):407–419. https://doi.org/10.1101/gr.119867.110
Goebel EA, Hernandez Bonilla S, Dong F, Dickson BC, Hoang LN, Hardisson D, Kolin DL (2019) Uterine tumor resembling ovarian sex cord tumor (UTROSCT): a morphologic and molecular study of 26 cases confirms recurrent NCOA1-3 rearrangement. Am J Surg Pathol. https://doi.org/10.1097/pas.0000000000001348(Epub ahead of print)
Gremel G, Liew M, Hamzei F, Hardell E, Selling J, Ghaderi M, Carlson JW (2015) A prognosis based classification of undifferentiated uterine sarcomas: identification of mitotic index, hormone receptors and YWHAE-FAM22 translocation status as predictors of survival. Int J Cancer 136(7):1608–1618. https://doi.org/10.1002/ijc.29141
Griewank KG, Wiesner T, Murali R, Pischler C, Muller H, Koelsche C, Sucker A (2018) Atypical fibroxanthoma and pleomorphic dermal sarcoma harbor frequent NOTCH1/2 and FAT1 mutations and similar DNA copy number alteration profiles. Mod Pathol 31(3):418–428. https://doi.org/10.1038/modpathol.2017.146
Hoang LN, Aneja A, Conlon N, DeLair D, Middha S, Benayed R, Chiang S (2017) Novel high-grade endometrial stromal sarcoma—a morphologic mimicker of myxoid leiomyosarcoma. Am J Surg Pathol 41(1):12–24
Hoang L, Chiang S, Lee CH (2018) Endometrial stromal sarcomas and related neoplasms: new developments and diagnostic considerations. Pathology 50(2):162–177. https://doi.org/10.1016/j.pathol.2017.11.086
Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, Northcott PA, Lichter P (2014) Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510(7506):537–541. https://doi.org/10.1038/nature13268
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Levine DA (2013) Integrated genomic characterization of endometrial carcinoma. Nature 497(7447):67–73. https://doi.org/10.1038/nature12113
Koelsche C, Hartmann W, Schrimpf D, Stichel D, Jabar S, Ranft A, von Deimling A (2018) Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol 31(8):1246–1256. https://doi.org/10.1038/s41379-018-0045-3
Koelsche C, Kriegsmann M, Kommoss FKF, Stichel D, Kriegsmann K, Vokuhl C, von Deimling A (2019) DNA methylation profiling distinguishes Ewing-like sarcoma with EWSR1-NFATc2 fusion from Ewing sarcoma. J Cancer Res Clin Oncol 145(5):1273–1281. https://doi.org/10.1007/s00432-019-02895-2
Kurman RJ, Carcangiu ML, Herrington CS, Young RH (2014) WHO classification of tumours of female reproductive organs. IARC, Lyon, p 6
Lewis N, Soslow RA, Delair DF, Park KJ, Murali R, Hollmann TJ, Chiang S (2018) ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol 31(4):674–684. https://doi.org/10.1038/modpathol.2017.162
Mariño-Enriquez A, Lauria A, Przybyl J, Ng L, Kowalewska M, Debiec-Rychter M, Fletcher J (2018) BCOR internal tandem duplication in high-grade uterine sarcomas. Am J Surg Pathol 42(3):335–341
McCluggage WG (2002) Uterine carcinosarcomas (malignant mixed Mullerian tumors) are metaplastic carcinomas. Int J Gynecol Cancer 12(6):687–690
Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Johann PJCC (2015) Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27(5):728–743. https://doi.org/10.1016/j.ccell.2015.04.002
Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, Davidson B, Micci F (2013) Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosom Cancer 52(7):610–618. https://doi.org/10.1002/gcc.22057
Rohrich M, Koelsche C, Schrimpf D, Capper D, Sahm F, Kratz A, Reuss DE (2016) Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol 131(6):877–887. https://doi.org/10.1007/s00401-016-1540-6
Sandoval J, Esteller M (2012) Cancer epigenomics: beyond genomics. Curr Opin Genet Dev 22(1):50–55. https://doi.org/10.1016/j.gde.2012.02.008
Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, Hoshino N (2015) Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 6:7558. https://doi.org/10.1038/ncomms8557
Shen H, Laird PW (2013) Interplay between the cancer genome and epigenome. Cell 153(1):38–55. https://doi.org/10.1016/j.cell.2013.03.008
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Staats PN, Garcia JJ, Dias-Santagata DC, Kuhlmann G, Stubbs H, McCluggage WG, Oliva E (2009) Uterine tumors resembling ovarian sex cord tumors (UTROSCT) lack the JAZF1–JJAZ1 translocation frequently seen in endometrial stromal tumors. Am J Surg Pathol 33(8):1206–1212. https://doi.org/10.1097/PAS.0b013e3181a7b9cf
Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Kool M (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164(5):1060–1072. https://doi.org/10.1016/j.cell.2016.01.015
Talhouk A, McConechy MK, Leung S, Li-Chang HH, Kwon JS, Melnyk N, McAlpine JN (2015) A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer 113(2):299–310. https://doi.org/10.1038/bjc.2015.190
van der Maaten L, Hinton G (2008) Visualizing Data using t-SNE. J Mach Learn Res 9:2579–2605
Acknowledgements
We want to thank the Microarray Unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ) for providing excellent technical support. This work was supported by the German Cancer Aid (Grant: 70112499). The laboratory of TGPG was supported by the Dr. Leopold and Carmen Ellinger foundation and the German Cancer Aid (Grant: 70112257).
Author information
Authors and Affiliations
Contributions
AvD, FKFK and CK conceptualized the project, FKFK and CK wrote the original draft. FKFK and CK coordinated data generation. CK, FS and AvD supervised RNA sequencing and DNA methylation array analysis, DSchr and DSt analyzed data. FKFK, MK, BT-C, AT, JNM, KTC, DS, SMP, LR-P, TK, TGPG, RB, HS, GM, PS, DS, HL, CBG, DGH, FK, AvD and CK provided tumor samples and corresponding metadata. All authors reviewed and approved of the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
PS: grants from MSD, BMS, Roche, Astra Zeneca, Novartis, Sanofi-Aventis, Chugai, Illumina and Thermo Fisher; personal fees from MSD, BMS, Roche, Astra Zeneca, AbbVie, Ipsen and Novartis. FS: lecturing honoraria from Illumina, Medac and Agilent; board member of AbbVie. SMP and AvD: patent pending for a “DNA-methylation based method for classifying tumors”. AvD: issued patent for IDH1R132H and BRAFv600E antibodies. All other authors declare no conflict of interest.
Ethical approval
This study was performed in accordance with the ethical standards of the institutional research committee and the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kommoss, F.K.F., Stichel, D., Schrimpf, D. et al. DNA methylation-based profiling of uterine neoplasms: a novel tool to improve gynecologic cancer diagnostics. J Cancer Res Clin Oncol 146, 97–104 (2020). https://doi.org/10.1007/s00432-019-03093-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-019-03093-w