Abstract
Purpose
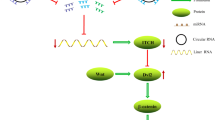
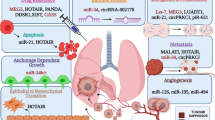
Circular RNAs (circRNAs), a large class of non-coding RNAs with covalently closed-loop structures, are abundant, stable, conserved, and have tissue and developmental-stage specificities. The biological functions of circRNAs are varied. Moreover, circRNAs participate in various pathological processes, especially in multiple cancers. Lung cancer is the most frequent malignant tumor worldwide. Many studies have suggested that circRNAs are pivotal in non-small cell lung cancer. This article aims to provide a retrospective review of the latest research on the functions of circRNAs in non-small cell lung cancer. In particular, we focus our discussion on the role of circRNAs in cell-cycle regulation and the epithelial–mesenchymal transition, and also discuss the known regulatory molecular mechanisms of circRNAs in non-small cell lung cancer.
Methods
We reviewed the literature on circRNAs and non-small cell lung cancer from PubMed databases. Specifically, we focused on the roles and mechanisms of circRNAs in regulating the cell cycle and the epithelial–mesenchymal transition.
Results
Dysregulation of circRNAs is closely correlated with proliferation, migration, and invasion of non-small cell lung cancer, especially in terms of modulating cell-cycle regulation and the epithelial–mesenchymal transition.
Conclusion
Taken together, circRNAs have potential as biomarkers for the diagnosis, prognosis, and treatment of non-small cell lung cancer.



Similar content being viewed by others
References
Abbas T, Dutta A (2009) p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9:400–414
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14:361–369
Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H (2015) Rolling circle translation of circular RNA in living human cells. Sci Rep 5:16435
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66
Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X (2015) The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 61:221–230
Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ (2014) Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33:981–993
Beermann J, Piccoli MT, Viereck J, Thum T (2016) Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 96:1297–1325
Chen CY, Sarnow P (1995) Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–417
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66:115–132
Chen D, Ma W, Ke Z, Xie F (2018) CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17:2080–2090
Chin L, Hahn WC, Getz G, Meyerson M (2011) Making sense of cancer genomic data. Genes Dev 25:534–555
Cho E-C, Kuo M-L, Liu X, Yang L, Hsieh Y-C, Wang J, Cheng Y, Yen Y (2014) Tumor suppressor FOXO3 regulates ribonucleotide reductase subunit RRM2B and impacts on survival of cancer patients. Oncotarget 5:4834–4844
Cocquerelle C, Mascrez B, Hétuin D, Bailleul B (1993) Mis-splicing yields circular RNA molecules. FASEB J 7:155–160
Collins LG, Haines C, Perkel R, Robert EE (2007) Lung cancer: diagnosis and management. Am Fam Physician 75:56–63
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134
Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z, Wang Z, Yang C, Jiang Y (2018) RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA 0006916, with important functions in lung cancer cells. Carcinogenesis 39(8):981–992
Ding L, Yao W, Lu J, Gong J, Zhang X (2018) Upregulation of circ_001569 predicts poor prognosis and promotes cell proliferation in non-small cell lung cancer by regulating the Wnt/beta-catenin pathway. Oncol Lett 16:453–458
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L (2016) CircRNA-derived pseudogenes. Cell Res 26:747–750
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–2858
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB (2017a) Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 24:357–370
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB (2017b) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412
Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, Liang H, Gao L (2019) A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548 g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett 442:222–232
Filbin ME, Kieft JS (2009) Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19:267–276
Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J (2017) Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget 8:58405
Gu X, Wang G, Shen H, Fei X (2018) Hsa_circ_0033155: a potential novel biomarker for non-small cell lung cancer. Exp Ther Med 16:3220–3226
Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP (2016) Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165:289–302
Guo T, Li J, Zhang L, Hou W, Wang R, Zhang J, Gao P (2019) Multidimensional communication of microRNAs and long non-coding RNAs in lung cancer. J Cancer Res Clin Oncol 145:31–48
Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, Cui J (2018) CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun 503:2429–2435
Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, Hu Z, Dai J, Shen H (2018) A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med 7:2783–2791
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Hochegger H, Takeda S, Hunt T (2008) Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol 9:910–916
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ (2017) Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77:2339
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10:170–177
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157
Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, Di QG (2018) Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol 144:667–674
Karreth FA, Pandolfi PP (2013) ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov 3:1113–1121
Kastan MB, Jiri B (2004) Cell-cycle checkpoints and cancer. Nature 432:316
Katayama R, Khan TM, Benes C, Lifshits E, Ebi H, Rivera VM, Shakespeare WC, Iafrate AJ, Engelman JA, Shaw AT (2011) Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci 108:7535–7540
Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H (1986) The hepatitis delta (delta) virus possesses a circular RNA. Nature 323:558–560
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Mark W (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22–37
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015a) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015b) Exon–intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264
Li J, Wang J, Chen Z, Chen Y, Jin M (2018a) Hsa_circ_0079530 promotes cell proliferation and invasion in non-small cell lung cancer. Gene 665:1–5
Li S, Sun X, Miao S, Lu T, Wang Y, Liu J, Jiao W (2018b) hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer 9:924–930
Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, Zhang S, Wang H, Qin W, Lu ZJ, Guo Y, Zhu Q, Wang D (2019) Microarray is an efficient tool for circRNA profiling. Brief Bioinform 20(4):1420–1433
Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X, Guan G, Dong X (2018d) Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun 503:2089–2094
Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL (2017) Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res 7:1566–1576
Liu T, Song Z, Gai Y (2018a) Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun 503:1503–1509
Liu W, Ma W, Yuan Y, Zhang Y, Sun S (2018b) Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun 500:846–851
Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW, Wen J (2017) Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother 96:892–898
Ma X, Yang X, Bao W, Li S, Liang S, Sun Y, Zhao Y, Wang J, Zhao C (2018) Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun 498:1009–1015
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA (2008) The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715
Massague J (2008) TGFbeta in cancer. Cell 134:215–230
Massagué J (2004) G1 cell-cycle control and cancer. Nature 432:298
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Memczak S, Papavasileiou P, Peters O, Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10:e0141214
Myatt SS, Lam EWF (2007) The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 7:847–859
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S (2017) Translation of CircRNAs. Mol Cell 66:9–21 (e7)
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D (2018) Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem 119:440–446
Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7:415–428
Qi Y, Zhang B, Wang J, Yao M (2018) Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene 676:79–85
Qin M, Wei G, Sun X (2018) Circ-UBR5: an exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem Biophys Res Commun 503:1027–1034
Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, Xu W, Zhang E, Wang J, Fang T, Hu J, Dong G, Yin R, Wang J, Xu L (2018) The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res 78:2839–2851
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:141–148
Qu D, Yan B, Xin R, Ma T (2018) A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res 8:1387–1402
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146:353–358
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73:3852–3856
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H (2007) Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448:561–566
Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W (2016) Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 44:e87
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A (2011) An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147:370–381
Sun C, Huang C, Li S, Yang C, Xi Y, Wang L, Zhang F, Fu Y, Li D (2016) Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget 7:8341–8359
Suzuki H, Tsukahara T (2014) A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 15:9331–9342
Tan SM, Kirchner R, Jin J, Hofmann O, McReynolds L, Hide W, Lieberman J (2014) Sequencing of captive target transcripts identifies the network of regulated genes and functions of primate-specific miR-522. Cell Rep 8:1225–1239
Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, Liu Y, Liu L, Wei YQ, Peng Y (2018a) Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res 28:693–695
Tan S, Sun D, Pu W, Gou Q, Guo C, Gong Y, Li J, Wei YQ, Liu L, Zhao Y, Peng Y (2018b) Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer 17:138
Tian F, Yu CT, Ye WD, Wang Q (2017) Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem Biophys Res Commun 493:1260–1266
Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W (2019) CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem 120:3765–3772
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65:87–108
Van Der Vos KE, Coffer PJ (2011) The extending network of FOXO transcriptional target genes. Antioxid Redox Signal 14:579–592
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ (2016) Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int 2016:1579490
Wang J, Li H (2018) CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci 22:3053–3060
Wang Y, Wang Z (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21:172–179
Wang L, Liu S, Mao Y, Xu J, Yang S, Shen H, Xu W, Fan W, Wang J (2018a) CircRNF13 regulates the invasion and metastasis in lung adenocarcinoma by targeting miR-93-5p. Gene 671:170–177
Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, Su Z, Zhang C, Liu X, Xu G, Zhang HT (2018b) Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial–mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer 17(1):140
Wang X, Zhu X, Zhang H, Wei S, Chen Y, Chen Y, Wang F, Fan X, Han S, Wu G (2018c) Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun 496:1069–1075
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A (2017) Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 23:3918
Wood DE, George AE, David SE, Hou L, Jackman D, Kazerooni E, Klippenstein D, Rudy PL, Leard L, Ann NCL (2012) Lung cancer screening. J Natl Comp Cancer Netw 10:240–265
Wu K, House L, Liu W, Cho WC (2012) Personalized targeted therapy for lung cancer. Int J Mol Sci 13:11471–11496
Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M (2017a) The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol 143:17–27
Xu T, Wu J, Han P, Zhao Z, Song X (2017b) Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genom 18:680
Yang W, Du WW, Li X, Yee AJ, Yang BB (2016) Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 35:3919–3931
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, Dong J, Khorshidi A, Yang BB (2017a) A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ 24:1609–1620
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z (2017b) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27:626–641
Yang L, Wang J, Fan Y, Yu K, Jiao B, Su X (2018) Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respir Res 19:115
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ (2017) Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract 213:453–456
Yilmaz M, Christofori G (2009) EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 28:15–33
Yu J, Xu Q, Wang Z, Yang Y, Zhang L, Ma J, Sun S, Yang F, Zhou W (2018a) Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 68:1214–1227
Yu W, Jiang H, Zhang H, Li J (2018b) Hsa_circ_0003998 promotes cell proliferation and invasion by targeting miR-326 in non-small cell lung cancer. Onco Targets Ther 11:5569–5577
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159:134–147
Zhang Y, Dai J, Deng H, Wan H, Liu M, Wang J, Li S, Li X, Tang H (2015) miR-1228 promotes the proliferation and metastasis of hepatoma cells through a p53 forward feedback loop. Br J Cancer 112:365–374
Zhang H, Yu C, Chen M, Li Z, Tian S, Jiang J, Sun C (2016) miR-522 contributes to cell proliferation of hepatocellular carcinoma by targeting DKK1 and SFRP2. Tumour Biol 37:11321–11329
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N (2018a) A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37:1805–1814
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H (2018b) Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep 8:2878
Zhang X, Yang D, Wei Y (2018c) Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther 11:3979–3987
Zhang Y, Zhao H, Zhang L (2018d) Identification of the tumorsuppressive function of circular RNA FOXO3 in nonsmall cell lung cancer through sponging miR155. Mol Med Rep 17:7692–7700
Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M (2017) CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem 44:2138–2146
Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K (2018) circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. https://doi.org/10.1042/BSR20180570
Zhou R, Chen KK, Zhang J, Xiao B, Huang Z, Ju C, Sun J, Zhang F, Lv XB, Huang G (2018) The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer 17:75
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, Han S, Wu G (2017) hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J 284:2170–2182
Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li D, Zhang L (2018) Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother 102:639–644
Zou Q, Wang T, Li B, Li G, Zhang L, Wang B, Sun S (2018) Overexpression of circ-0067934 is associated with increased cellular proliferation and the prognosis of non-small cell lung cancer. Oncol Lett 16:5551–5556
Acknowledgements
All of the data generated or analyzed during this study are included in this published article.
Funding
Funding was provided by the National Natural Science Foundation of China (Grant no. 81672297).
Author information
Authors and Affiliations
Contributions
JZ and JL contributed to the conception of the study. LZ, GM, QW, and XL contributed significantly to the analysis of data and preparation of the manuscript. CL performed data analyses and wrote the manuscript. All of the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, C., Zhang, L., Meng, G. et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol 145, 2875–2889 (2019). https://doi.org/10.1007/s00432-019-03045-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-019-03045-4