Abstract
Purpose
Despite considerable evidence that supports the NF-kB role in the immune system and lymphomagenesis, it is unclear whether specific NF-kB dimers control a particular set of genes that account for their biological functions. Our previous work showed that Hodgkin Lymphoma (HL) is unique, among germinal center (GC)-derived lymphomas, with respect to its dependency on Rel-B to survive. In contrast, diffuse large B-Cell lymphoma (DLBCL) including both Activated B-Cell-Like and Germinal Center B-Cell-Like, requires cREL and Rel-A to survive and it is not affected by Rel-B depletion. These findings highlighted the activity of specific NF-kB subunits in different GC-derived lymphomas.
Methods
Sequenced chromatin immunoprecipitated DNA fragments (ChIP-Seq) analysis revealed an extensive NF-kB DNA-binding network in DLBCL and HL. The ChIP-Seq data was merged with microarray analysis following the Rel-A, Rel-B or cRel knockdown to determine effectively regulated genes.
Results
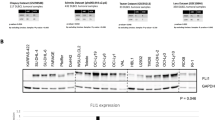
Downstream target analysis showed enrichment for cell cycle control, among other signatures. Rel-B and cRel controlled different genes within the same signature in HL and DLBCL, respectively. BCL2 was exclusively controlled by Rel-B in HL. Both mRNA and protein levels decreased following Rel-B depletion meanwhile there was no change upon cRel knock-down. BCL2 exogenous expression partially rescued the death induced by decreased Rel-B in HL cells.
Conclusion
The Rel-B hierarchical network defined HL and the cRel hierarchical network characterized DLBCL. Each Rel member performs specific functions in distinct GC-derived lymphomas. This result should be considered for the development of targeted therapies that are aimed to selectively inhibit individual NF-kB dimers.




Similar content being viewed by others
Data availability
Availability of data and materials:Expression arrays data have been deposited in the NCBI GEO database under the GSE109803 accession number. ChIP-Seq data have been deposited in the NCBI GEO database under the GSE110348 accession number and can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110348. All gene lists that are derived from our analysis of the ChIP-Seq and expression arrays experiments, as well as more detailed protocols, are available upon request.
References
Alizadeh AA, Eisen MB, Davis RE et al (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403(6769):503–511. https://doi.org/10.1038/35000501
Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Melzier G, Vedenko A et al (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324(5935):1720–1723. https://doi.org/10.1126/science.1162327
Barth TF, Martín-Subero JI, Joos S, Menz CK, Hasel C, Mechtersheimer G, Parwaresch RM, Lichter P, Siebert R et al (2003) Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin Lymphoma. Blood 101:3681–3686. https://doi.org/10.1182/blood-2002-08-2577
Cabannes E, Khan G, Aillet F, Jarrett RE, Hay RT (1999) Mutations in the IkBa gene in Hodgkin’s disease suggest a tumour suppressor role for IkappaBalpha. Oncogene 18(20):3063–3070. https://doi.org/10.1038/sj.onc.1202893
Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES (2011) The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 117:5019–5032
Carbone A, Gloghini A, Cabras A, Elia G (2009) The Germinal centre-derived lymphomas seen through their cellular microenvironment. Br J Haematol 145(4):468–480. https://doi.org/10.1111/j.1365-2141.2009.07651.x
Compagno M, Lim WK, Grunn A et al (2009) Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 459(7247):717–721. https://doi.org/10.1038/nature07968
Davis RE, Brown KD, Siebenlist U, Staudt LM (2001) Constitutive nuclear factor kappa B activity is required for survival of activated B Cell-like diffuse large B cell lymphoma cells. J Exp Med 194:1861–1874. https://doi.org/10.1038/nature07968
Davis RE, Ngo VN, Lenz G, Tolar P, Young Y, Romesser PB, Kohlhammer H, Lamy L, Zhao H et al (2010) Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463(7277):88–92. https://doi.org/10.1038/nature08638
De la Paz NG, Simeonidis S, Leo C, Rose DW, Collins T (2007) Regulation of NF-kappaB-dependent gene expression by the POU domain trasncription factor. J Biol Chem 282(1):8424–8434. https://doi.org/10.1074/jbc.M606923200
Emmerich F, Meiser M, Hummel M et al (1999) Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood 94(9):3129–3134
Emmerich F, Theurich S, Hummel M et al (2003) Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J Pathol 201(3):413–420. https://doi.org/10.1002/path.1454
Feuerhake F, Kutok JL, Monti S, Chen W, LaCasce AS, Cattoretti G, Kurlin P, Pinkus GS, de Leval L et al (2005) NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood 106:1392–1399. https://doi.org/10.1182/blood-2004-12-4901
Gilmore TD, KalaitzidisD Liang MC, Starczynowski DT (2004) The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene 23(13):2275–2286. https://doi.org/10.1038/sj.onc.1207410
Harris NL, Jaffe ES, Diebold J et al (1997) The World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. Report of the Clinical Advisory Committee meeting, Airlie House, Virginia. Ann Oncol 10(12):1419–1432
Joos S, Menz CK, Wrobel G et al (2002) Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome. Blood 99(4):1381–1387
Jungnicket B, Staratschek-Jox A, Brauninger A et al (2000) Clonal deleterious mutations in the IkBa gene in the malignant cells in Hodgkin’s disease. J Exp Med 191:395–401
Kapatai G, Murray P (1999) Contribution of the Epstein Barr virus to the molecular pathogenesis of Hodgkin lymphoma. J Clin Pathol 60(12):1342–1349. https://doi.org/10.1136/jcp.2007.050146
Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S et al (2009) Frequent inactivation of A20 in B-cell lymphomas. Nature 459(7247):712–716. https://doi.org/10.1038/nature07969
Kerr LD, Ransone LJ, Wamsley P, Schmitt MJ, Bover TG, Zhou Q, Berk AJ, Verma IM (1993) Association between proto-oncoprotein Rel and TATA-binding protein mediates transcriptional activation by NF-kappa B. Nature 365(6445):4
Klein U, Heise N (2015) Unexpected functions of NF-kB during germinal center B-Cell development: implications for lymphomagenesis. Curr Opin Hematol 22(4):379–387. https://doi.org/10.1097/moh.0000000000000160
Klein U, Tu Y, Stolovitzky GA et al (2003) Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci USA 100(5):2639–2644. https://doi.org/10.1073/pnas.0437996100
Kondo E, Yoshino T (2007) Expression of apoptosis regulators in germinal centers and germinal center-derived B-cell lymphomas: insight into B-cell lymphomagenesis. Pathol Int 57(7):391–397. https://doi.org/10.1111/j.1440-1827.2007.02115.x
Küppers R (2009) The biology of Hodgkin’s Lymphoma. Nat Rev Cancer Nat 459(7247):712–716. https://doi.org/10.1038/nrc2542
Küppers R, Rajewiski K (1998) The origin of Hodgkin and Reed/Sternberg cells in Hodgkin disease. Annu Rev Immunol 16:471–493. https://doi.org/10.1146/annurev.immunol.16.1.471
Lake A, Shield LA, Cordano P et al (2009) Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. Int J Cancer 125(6):1334–1342. https://doi.org/10.1002/ijc.24502
Lenz G, Davis RE, Ngo VN, Lloyd L, George TC, Wright GW, Dave SS, Zhao W, Xu W et al (2008) Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319(5870):1676–1679. https://doi.org/10.1126/science.1153629
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Martin-Subero JI, Gesk S, Harder L, Sonoki T, Tucker PW, Schlegelberger B, Grote W, Novp FJ, Calasanz MJ et al (2002) Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood 99:1474–1477
Milanovic M, Heise N, De Silva NS, Anderson MM, Silva K, Carette A, Orelli F, Bhagat G, Klein U et al (2017) Differential requirements for the canonical NF-kB transcription factors c-REL and REL-A in the generation and maturation of mature B cells. Immunol Cell Biol 95(3):261–271. https://doi.org/10.1084/jem.20132613
Ngo VN, Davis RE, Lamy L et al (2006) A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441(7089):106–110. https://doi.org/10.1038/nature04687
Oeckinghaus A, Ghosh S (2010) The NF-kB family of transcription factors and its regulation. Cold Spring Harbor Laboratory Press, pp 5–18. https://doi.org/10.1101/cshperspect.a000034
Ranuncolo SM, Pittaluga S, Evbuomwan MO, Jaffe ES, Lewis BA (2012) Hodgkin Lymphoma requires stabilized NIK and constitutive relB expression for survival. Blood 120(18):3756–3763. https://doi.org/10.1182/blood-2012-01-405951
Saitoh Y, Yamamoto N, Dewan MZ, Sugimoto H, Martinez Bruyn VJ, Iwasaki Y, Matsubara K, Qi X, Saitoh T et al (2008) Overexpressed NF-kappaB-inducing kinase contributes to the tumorigenesis of adult T-cell leukemia and Hodgkin Reed-Sternberg cells. Blood 111(10):5118–5129. https://doi.org/10.1182/blood-2007-09-110635
Schmitz R, Hansmann ML, Bohle V et al (2009a) TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 206(5):981–989. https://doi.org/10.1084/jem.20090528
Schmitz R, Stanelle J, Hansmann MS, Küppers R (2009b) Pathogenesis of classical and lymphocyte-predominant Hodgkin lymphoma. Annu Rev Pathol 4:151–174. https://doi.org/10.1146/annurev.pathol.4.110807.092209
Shih VF, Tsui R, Caldwell A, Hoffmann A (2011) A single NFkB system for both canonical and non-canonical signaling. Cell Res 21(1):86–102. https://doi.org/10.1038/cr.2010.161
Sun S-C (2011) Non-canonical NF-kB signaling pathway. Cell Res 21:71–78. https://doi.org/10.1038/cr.2010.177
Thome M, Charton JE, Pelzer C, Hailfinger S (2010) Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol 9:a003004. https://doi.org/10.1101/cshperspect.a003004
Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, Lira SA, Bravo R (1995) Multiorgan inflammation and hematopoietic abnormalities in mice with targeted disruption of RelB, a member of the NF-kB/Rel family. Cell 80:331–340 PMID: 7834753
Weih DS, Yilmaz ZB, Weih F (2001) Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J Immunol 167:1909–1919 PMID: 11489970
WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. WHO classification of tumors (2008) Revised 4th Edition Volume 2. Edited by Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. Lyon: IARC Press 323–325
Wong D, Teixeira A, Oikonomopoulos S, Humburg P, Lone IN, Saliba D, Siggers T, Bulyk M, Angelov D et al (2011) Extensive characterization of NF-kB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biology 12:R70. https://doi.org/10.1186/gb-2011-12-7-r70
Zarnegar B, Yamazaki S, He JQ, Cheng G (2011) Control of canonical NF-kB activation through the NIK-IKK complex pathway. Edited by Owen N. Witte, University of California, Los Angeles, CA vol. 105 N 9:3503-3508. https://doi.org/10.1073/pnas.0707959105
Funding
Instituto Universitario del Hospital Italiano (IUHI), Buenos Aires, Argentina. Fundación para el Progreso de la Medicina (FPM), Ciudad de Córdoba, Córdoba, Argentina. Intramural Program of the National Cancer Institute (NCI), National Institutes of Health (NIH).
Author information
Authors and Affiliations
Contributions
AMGC and MC performed experiments, WX analyzed the ChIP-Seq data and developed the algorithm for this purpose. TAW discussed results. SMR performed experiments, analyzed data, discussed results and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gamboa-Cedeño, A.M., Castillo, M., Xiao, W. et al. Alternative and canonical NF-kB pathways DNA-binding hierarchies networks define Hodgkin lymphoma and Non-Hodgkin diffuse large B Cell lymphoma respectively. J Cancer Res Clin Oncol 145, 1437–1448 (2019). https://doi.org/10.1007/s00432-019-02909-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-019-02909-z