Abstract
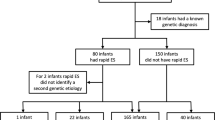
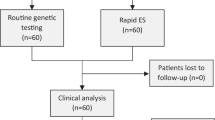
Genetic disorders are one of the leading causes of infant mortality and are frequent in neonatal intensive care units (NICUs). Rapid genome-wide sequencing (GWS; whole genome or exome sequencing (ES)), due to its diagnostic capabilities and immediate impacts on medical management, is becoming an appealing testing option in the NICU setting. RAPIDOMICS was a trio-based rapid ES pilot study of 25 babies with suspected genetic disorders in the BC Women’s Hospital NICU. ES and bioinformatic analysis were performed after careful patient ascertainment. Trio analysis was performed using an in-house pipeline reporting variants in known disease-causing genes. Variants interpreted by the research team as definitely or possibly causal of the infant’s phenotype were Sanger validated in a clinical laboratory. The average time to preliminary diagnosis was 7.2 days. Sanger validation was pursued in 15 patients for 13 autosomal dominant and 2 autosomal recessive disorders, with an overall diagnostic rate (partial or complete) of 60%.
Conclusion: In total, 72% of patients enrolled had a genomic diagnosis achieved through ES, multi-gene panel testing or chromosomal microarray analysis. Among these, there was an 83% rate of significant and immediate impact on medical decision-making directly related to new knowledge of the diagnosis. Health service implementation challenges and successes are discussed.
What is Known: • Rapid genome-wide sequencing in the neonatal intensive care setting has a greater diagnostic hit rate and impact on medical management than conventional genetic testing. However, the impact of consultation with genetics and patient ascertainment requires further investigation. | |
What is New: • This study demonstrates the importance of genetic consultation and careful patient selection prior to pursuing exome sequencing (ES). • In total, 15/25 (60%) patients achieved a diagnosis through ES and 18/25 (72%) through ES, multi-gene panel testing or chromosomal microarray analysis with 83% of those having immediate effects on medical management. |
Similar content being viewed by others
Abbreviations
- NICU:
-
Neonatal intensive care unit
- GWS:
-
Genome-wide sequencing
- ES:
-
Exome sequencing
- WGS:
-
Whole genome sequencing
- OMIM:
-
Online Mendelian Inheritance in Man
- CMA:
-
Chromosomal microarray analysis
- CMV:
-
Cytomegalovirus
- TPN:
-
Total parenteral nutrition
- VUS:
-
Variant of uncertain significance
References
Weiner J, Sharma J, Lantos J, Kilbride H (2011) How infants die in the neonatal intensive care unit: trends from 1999 through 2008. Arch Pediatr Adolesc Med 165(7):630–634. https://doi.org/10.1001/archpediatrics.2011.102
Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, Zhang J, He W, Dharmadhikari AV, Qu C, Ward P, Braxton A, Narayanan S, Ge X, Tokita MJ, Santiago-Sim T, Dai H, Chiang T, Smith H, Azamian MS, Robak L, Bostwick BL, Schaaf CP, Potocki L, Scaglia F, Bacino CA, Hanchard NA, Wangler MF, Scott D, Brown C, Hu J, Belmont JW, Burrage LC, Graham BH, Sutton VR, Craigen WJ, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Muzny DM, Miller MJ, Wang X, Leduc MS, Xiao R, Liu P, Shaw C, Walkiewicz M, Bi W, Xia F, Lee B, Eng CM, Yang Y, Lalani SR (2017) Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr 171(12):e173438. https://doi.org/10.1001/jamapediatrics.2017.3438
Bowdin SC (2016) The clinical utility of next-generation sequencing in the neonatal intensive care unit. CMAJ. 188(11):786–787. https://doi.org/10.1503/cmaj.160490
Stark Z, Lunke S, Brett GR et al (2018) Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med 20:1554–1563. https://doi.org/10.1038/gim.2018.37
van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, de Koning TJ, Sikkema-Raddatz B, van der Velde JK, Abbott KM, Herkert JC, Löhner K, Rump P, Meems-Veldhuis MT, Neerincx PBT, Jongbloed JDH, van Ravenswaaij-Arts CM, Swertz MA, Sinke RJ, van Langen IM, Wijmenga C (2017) Rapid targeted genomics in critically ill newborns. Pediatrics. 140(4):e20162854. https://doi.org/10.1542/peds.2016-2854
Willig L, Petrikin J, Smith L et al (2015) Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med 3(5):377–387. https://doi.org/10.1016/S2213-2600(15)00139-3
Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, Saunders CJ, Thiffault I, Miller NA, Zellmer L, Herd SM, Holmes AM, Batalov S, Veeraraghavan N, Smith LD, Dimmock DP, Leeder JS, Kingsmore SF (2018) The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genomic Med 3(1):6. https://doi.org/10.1038/s41525-018-0045-8
Berg JS, Agrawal PB, Bailey DB et al (2017) Newborn sequencing in genomic medicine and public health. Pediatrics. 139(2):e20162252. https://doi.org/10.1542/peds.2016-2252
Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF (2012) Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 4(154):154ra135. https://doi.org/10.1126/scitranslmed.3004041
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG (2009) Research electronic data capture (REDCap) - a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42(2):377–381. https://doi.org/10.1016/j.jbi.2008.08.010
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. https://doi.org/10.1093/nar/gkq603
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20(1):110–121. https://doi.org/10.1101/gr.097857.109
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4(7):1073–1081. https://doi.org/10.1038/nprot.2009.86
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. https://doi.org/10.1038/nmeth0410-248
Chun S, Fay JC (2009) Identification of deleterious mutations within three human genomes. Genome Res 19(9):1553–1561. https://doi.org/10.1101/gr.092619.109
Schwarz JM, Rödelsperger C, Schuelke M, Seelow D (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7(8):575–576. https://doi.org/10.1038/nmeth0810-575
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46(3):310–315. https://doi.org/10.1038/ng.2892
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(D1):D862–D868. https://doi.org/10.1093/nar/gkv1222
Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LELM, Wissink-Lindhout W, Fenckova M, van den Akker WMR, Kasri NN, Nillesen WM, Prescott T, Clark RD, Devriendt K, van Reeuwijk J, de Brouwer APM, Gilissen C, Zhou H, Brunner HG, Veltman JA, Schenck A, van Bokhoven H (2012) Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet 91(1):73–82. https://doi.org/10.1016/j.ajhg.2012.05.003
Sharma D, Kumar C, Bhalerao S, Pandita A, Shastri S, Sharma P (2015) Ectrodactyly, ectodermal dysplasia, cleft lip, and palate (EEC syndrome) with tetralogy of fallot: a very rare combination. Front Pediatr 3:51. https://doi.org/10.3389/fped.2015.00051
Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, Marrs T, Corder S, Krivohlavek L, Walter A, Petrikin JE, Saunders CJ, Thiffault I, Soden SE, Smith LD, Dinwiddie DL, Herd S, Cakici JA, Catreux S, Ruehle M, Kingsmore SF (2015) A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med 7(1):100. https://doi.org/10.1186/s13073-015-0221-8
Elliott AM, du Souich C, Adam S, Dragojlovic N, van Karnebeek C, Nelson TN, Lehman A, The CAUSES Study, Lynd LD, Friedman JM (2018) The genomic consultation service: a clinical service designed to improve patient selection for genome-wide sequencing in British Columbia. Mol Genet Genomic Med 6:592–600. https://doi.org/10.1002/mgg3.410
Smith E, du Souich C, Dragojlovic N, CAUSES Study, RAPIDOMICS Study, Elliott AM (2019) Genetic counseling considerations with rapid genome-wide sequencing in a neonatal intensive care unit. J Genet Couns 28(2):263–272. https://doi.org/10.1002/jgc4.1074
Acknowledgments
We thank the families who participated in this study, Jennifer Claydon, and clinical staff who assisted with coordination. We are grateful to Drs. Susan Albersheim, Michael Castaldo, Deepak Manhas, Julia Panczuk, Jonathan Wong, and Grace Yu from Neonatology who contributed to patient selection and follow-up. RAPIDOMICS study investigators include the following: Tara Candido, Jan Christilaw, Nick Dragojlovic, Christèle du Souich, Alison M. Elliott, Daniel M. Evans, Matthew J. Farrer, Jan M. Friedman, Ilaria Guella, Anna Lehman, Larry Lynd, Horacio Osiovich, and Leah Tooman.
Funding
This project was funded by the User Partnership Program (UPP): Genome British Columbia with co-funding from the Provincial Health Services Authority (PHSA). The Centre for Applied Neurogenetics was supported by the Canada Excellence Research Chairs program (MJF). Leading Edge Endowment Funds from the Province of British Columbia, LifeLabs, and Genome BC support the Dr. Donald Rix BC Leadership Chair (MJF). “Neuroseq” bioinformatics development is supported through the Canadian Consortium on Neurodegeneration in Aging and the Pacific Parkinson’s Research Institute.
Author information
Authors and Affiliations
Contributions
Dr. Elliott conceptualized and designed the study; was involved in data acquisition, analysis, and interpretation; drafted the initial manuscript; and revised the manuscript.
Ms. du Souich, Dr. Lehman, and Dr. Horacio Osiovich conceptualized and designed the study, were involved in data acquisition and analysis and interpretation of the data, reviewed and revised the manuscript.
Dr. Friedman conceptualized and designed the study, was involved in analysis and interpretation of the data, and reviewed and critically revised the manuscript.
Dr. Guella, Mr. Evans, Ms. Candido, Ms. Tooman, and Dr. Farrer conceptualized and designed the study, were involved in data analysis and interpretation, and reviewed and revised the manuscript.
Drs. Armstrong, Clarke, Gibson, Gill, Lewis, McKinnon, Nikkel, Patel, and van Allen were involved in data collection and interpretation of the data and critically revised the manuscript.
Drs. Lavoie, Solimano, Synnes, and Ting were involved in data collection and interpretation and critically revised the manuscript.
Dr. Christilaw conceptualized and designed the study, facilitated data acquisition, and provided revision of the manuscript.
All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the University of British Columbia (UBC)/BC Children’s and Women’s Clinical Research Ethics Board (REB H15-02750).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by Patrick Van Reempts
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Revisions received: 4 May 2019 / 7 May 2019
Rights and permissions
About this article
Cite this article
Elliott, A.M., du Souich, C., Lehman, A. et al. RAPIDOMICS: rapid genome-wide sequencing in a neonatal intensive care unit—successes and challenges. Eur J Pediatr 178, 1207–1218 (2019). https://doi.org/10.1007/s00431-019-03399-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00431-019-03399-4