Abstract
Human immunodeficiency virus-1 tropism highly correlates with the amino acid (aa) composition of the third hypervariable region (V3) of gp120. A shift towards more positively charged aa is seen when binding to CXCR4 compared with CCR5 (X4 vs. R5 strains), especially positions 11 and 25 (11/25-rule) predicting X4 viruses in the presence of positively charged residues. At nucleotide levels, negatively or uncharged aa, e.g., aspartic and glutamic acid and glycine, which are encoded by the triplets GAN (guanine-adenosine-any nucleotide) or GGN are found more often in R5 strains. Positively charged aa such as arginine and lysine encoded by AAR or AGR (CGN) (R means A or G) are seen more frequently in X4 strains suggesting our hypothesis that a switch from R5 to X4 strains occurs via a G-to-A mutation. 1527 V3 sequences from three independent data sets of X4 and R5 strains were analysed with respect to their triplet composition. A higher number of G-containing triplets was found in R5 viruses, whereas X4 strains displayed a higher content of A-comprising triplets. These findings also support our hypothesis that G-to-A mutations are leading to the co-receptor switch from R5 to X4 strains. Causative agents for G-to-A mutations are the deaminases APOBEC3F and APOBEC3G. We therefore hypothesize that these proteins are one driving force facilitating the appearance of X4 variants. G-to-A mutations can lead to a switch from negatively to positively charged aa and a respective alteration of the net charge of gp120 resulting in a change of co-receptor usage.




Similar content being viewed by others
References
Berger EA, Murphy PM, Farber JM (1999) Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 17:657–700
Sierra S, Kaiser R, Thielen A, Lengauer T (2007) Genotypic coreceptor analysis. Eur J Med Res 12(9):453–462
Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI (1999) Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73(12):10489–10502
Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M (1992) Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol 66(3):1354–1360
Moyle GJ, Wildfire A, Mandalia S, Mayer H, Goodrich J, Whitcomb J, Gazzard BG (2005) Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis 191(6):866–872. doi:10.1086/428096
Bozzette SA, McCutchan JA, Spector SA, Wright B, Richman DD (1993) A cross-sectional comparison of persons with syncytium- and non-syncytium-inducing human immunodeficiency virus. J Infect Dis 168(6):1374–1379
Alimonti JB, Ball TB, Fowke KR (2003) Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J Gen Virol 84(Pt 7):1649–1661
Rosenberg YJ, Anderson AO, Pabst R (1998) HIV-induced decline in blood CD4/CD8 ratios: viral killing or altered lymphocyte trafficking? Immunol Today 19(1):10–17
Herbein G, Mahlknecht U, Batliwalla F, Gregersen P, Pappas T, Butler J, O’Brien WA, Verdin E (1998) Apoptosis of CD8+ T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature 395(6698):189–194
Zhang SY, Zhang Z, Fu JL, Kang FB, Xu XS, Nie WM, Zhou CB, Zhao M, Wang FS (2009) Progressive CD127 down-regulation correlates with increased apoptosis of CD8 T cells during chronic HIV-1 infection. Eur J Immunol 39(5):1425–1434
Delobel P, Nugeyre MT, Cazabat M, Pasquier C, Marchou B, Massip P, Barre-Sinoussi F, Israel N, Izopet J (2007) Population-based sequencing of the V3 region of env for predicting the coreceptor usage of human immunodeficiency virus type 1 quasispecies. J Clin Microbiol 45(5):1572–1580
De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J (1992) Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol 66(11):6777–6780
Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R (2007) Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 25(12):1407–1410
Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Sandres-Saune K, Cuzin L, Marchou B, Massip P, Izopet J (2008) Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. Aids 22(14):F11–F16
Pathak VK, Temin HM (1990) Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA 87(16):6019–6023
Pathak VK, Temin HM (1990) Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA 87(16):6024–6028
Preston BD, Poiesz BJ, Loeb LA (1988) Fidelity of HIV-1 reverse transcriptase. Science 242(4882):1168–1171
Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH (2004) Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14(15):1392–1396
Bishop KN, Holmes RK, Sheehy AM, Malim MH (2004) APOBEC-mediated editing of viral RNA. Science 305(5684):645
Sheehy AM, Gaddis NC, Choi JD, Malim MH (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418(6898):646–650
Wiegand HL, Doehle BP, Bogerd HP, Cullen BR (2004) A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 23(12):2451–2458
Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113(6):803–809
Lecossier D, Bouchonnet F, Clavel F, Hance AJ (2003) Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300(5622):1112
Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D (2003) Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424(6944):99–103
Liddament MT, Brown WL, Schumacher AJ, Harris RS (2004) APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol 14(15):1385–1391
Pace C, Keller J, Nolan D, James I, Gaudieri S, Moore C, Mallal S (2006) Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol 80(18):9259–9269
Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD (2005) Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog 1(1):e6
Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L (2003) The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424(6944):94–98
Nowarski R, Britan-Rosich E, Shiloach T, Kotler M (2008) Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol 15(10):1059–1066
Mulder LC, Harari A, Simon V (2008) Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci USA 105(14):5501–5506
Pillai SK, Wong JK, Barbour JD (2008) Turning up the volume on mutational pressure: is more of a good thing always better? (a case study of HIV-1 Vif and APOBEC3). Retrovirology 5:26
Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C (2009) HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog 5(5):e1000414
Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC (2005) Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435(7038):108–114
Chelico L, Pham P, Calabrese P, Goodman MF (2006) APOBEC3G DNA deaminase acts processively 3′–>5′ on single-stranded DNA. Nat Struct Mol Biol 13(5):392–399
Pion M, Granelli-Piperno A, Mangeat B, Stalder R, Correa R, Steinman RM, Piguet V (2006) APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J Exp Med 203(13):2887–2893
Kreisberg JF, Yonemoto W, Greene WC (2006) Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med 203(4):865–870
Trapp S, Derby NR, Singer R, Shaw A, Williams VG, Turville SG, Bess JW Jr, Lifson JD, Robbiani M (2009) Double-stranded RNA analog poly(I:C) inhibits human immunodeficiency virus amplification in dendritic cells via type I interferon-mediated activation of APOBEC3G. J Virol 83(2):884–895
Pido-Lopez J, Whittall T, Wang Y, Bergmeier LA, Babaahmady K, Singh M, Lehner T (2007) Stimulation of cell surface CCR5 and CD40 molecules by their ligands or by HSP70 up-regulates APOBEC3G expression in CD4(+) T cells and dendritic cells. J Immunol 178(3):1671–1679
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Fouchier RA, Groenink M, Kootstra NA, Tersmette M, Huisman HG, Miedema F, Schuitemaker H (1992) Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol 66(5):3183–3187
Briggs DR, Tuttle DL, Sleasman JW, Goodenow MM (2000) Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). Aids 14(18):2937–2939
Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E (2009) Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 5(6):e1000495
Han Y, Wang X, Dang Y, Zheng YH (2008) APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS Pathog 4(7):e1000095
Acknowledgments
The authors would like to thank all members of the laboratory, especially Finja Schweitzer, for critical discussions throughout the study. The professional technical assistance of Dörte Hammerschmidt and Monika Timmen-Wego is gratefully acknowledged. Additional thanks goes to the HIV-GRADE consortium, especially to Mark Oette, Krankenhaus der Augustinerinnen, Cologne; Gerd Fätkenheuer, University Hospital of Cologne, Cologne; Stefan Esser, University of Duisburg-Essen, Essen; Stefan Reuter, University of Düsseldorf, Düsseldorf; all Germany for providing the clinical data set. This work was supported by German Bundesministerium für Gesundheit grant number 310/4476-02/3, the German Bundesministerium für Bildung und Forschung grant number 01ES0712 and grant number 0315480C, and the European Commission Health project Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN) grant number 223131.
Author information
Authors and Affiliations
Corresponding author
Additional information
E. Heger and A. Thielen contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
430_2011_199_MOESM2_ESM.doc
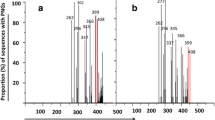
Supplementary Figure 1: Triplet and amino acid (aa) distribution in R5 and X4 strains. V3 sequences with defined phenotypes from the Los Alamos Database were analysed. According to the amino acid alignment, nucleotides were aligned and compared for their appearance in R5 (black) and X4 (grey). The percentage of each triplet (adenosine (abbreviated A), cytosine (C), guanine (G) and thymine (T) and the corresponding amino acid (Ala, alanine; Arg, arginine; Asn, aspargine; Asp, aspartic acid; Gln, glutamine; Glu, glutamic acid; Gly, glycine; Lys, lysine; Ser, serine; Thr, threonine, supplementary Table 1) for the position 11, 22, 24, and 25 are shown. Amino acids are grouped with respect to their physicochemical characteristics. (DOC 187 kb)
430_2011_199_MOESM3_ESM.doc
Supplementary Figure 2: G-to-A changes from R5 to X4 strains show highest correlation. The differences at each position in V3 between R5 and X4 strains regarding the percentage of nucleotide pairs (adenosine (abbreviated A), cytosine (C), guanine (G) and thymine (T) from the HIV-GRADE (A) and the Los Alamos (B) data set were calculated and correlated. Based on this calculation, the positions with the highest differences in R5 vs. X4 that mutate from one nucleotide to the other are located in the fourth (lower right) quadrant. The greatest correlation is boxed in red and enlarged (x ≥ 0.05, y ≤ -0.03). Nucleotide and triplet positions are indicated and separated by a slash. (DOC 1289 kb)
Rights and permissions
About this article
Cite this article
Heger, E., Thielen, A., Gilles, R. et al. APOBEC3G/F as one possible driving force for co-receptor switch of the human immunodeficiency virus-1. Med Microbiol Immunol 201, 7–16 (2012). https://doi.org/10.1007/s00430-011-0199-9
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-011-0199-9