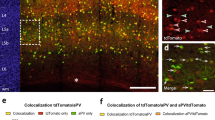
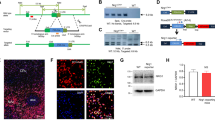
Abstract
In the present study, we generated a novel parvalbumin (PV)-Cre rat model and conducted detailed morphological and electrophysiological investigations of axons from PV neurons in globus pallidus (GP). The GP is considered as a relay nucleus in the indirect pathway of the basal ganglia (BG). Previous studies have used molecular profiling and projection patterns to demonstrate cellular heterogeneity in the GP; for example, PV-expressing neurons are known to comprise approximately 50% of GP neurons and represent majority of prototypic neurons that project to the subthalamic nucleus and/or output nuclei of BG, entopeduncular nucleus and substantia nigra (SN). The present study aimed to identify the characteristic projection patterns of PV neurons in the GP (PV-GP neurons) and determine whether these neurons target dopaminergic or GABAergic neurons in SN pars compacta (SNc) or reticulata (SNr), respectively. We initially found that (1) 57% of PV neurons co-expressed Lim-homeobox 6, (2) the PV-GP terminals were preferentially distributed in the ventral part of dorsal tier of SNc, (3) PV-GP neurons formed basket-like appositions with the somata of tyrosine hydroxylase, PV, calretinin and cholecystokinin immunoreactive neurons in the SN, and (4) in vitro whole-cell recording during optogenetic photo-stimulation of PV-GP terminals in SNc demonstrated that PV-GP neurons strongly inhibited dopamine neurons via GABAA receptors. These results suggest that dopamine neurons receive direct focal inputs from PV-GP prototypic neurons. The identification of high-contrast inhibitory systems on dopamine neurons might represent a key step toward understanding the BG function.









Similar content being viewed by others
References
Abdi A, Mallet N, Mohamed FY et al (2015) Prototypic and arkypallidal neurons in the dopamine-intact external globus pallidus. J Neurosci 35:6667–6688. doi:10.1523/JNEUROSCI.4662-14.2015
Atasoy D, Aponte Y, Su HH, Sternson SM (2008) A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci 28:7025–7030. doi:10.1523/JNEUROSCI.1954-08.2008
Bergman H, Feingold A, Nini A et al (1998) Physiological aspects of information processing in the basal ganglia of normal and parkinsonian primates. Trends Neurosci 21:32–38
Bevan MD, Smith AD, Bolam JP (1996) The substantia nigra as a site of synaptic integration of functionally diverse information arising from the ventral pallidum and the globus pallidus in the rat. Neuroscience 75:5–12. doi:10.1016/0306-4522(96)00377-6
Bevan MD, Booth PA, Eaton SA, Bolam JP (1998) Selective innervation of neostriatal interneurons by a subclass of neuron in the globus pallidus of the rat. J Neurosci 18:9438–9452
Bevan MD, Magill PJ, Terman D et al (2002) Move to the rhythm: oscillations in the subthalamic nucleus-external globus pallidus network. Trends Neurosci 25:525–531
Bolam JP, Smith Y (1990) The GABA and substance P input to dopaminergic neurones in the substantia nigra of the rat. Brain Res 529:57–78. doi:10.1016/0006-8993(90)90811-O
Carter DA, Fibiger HC (1978) The projections of the entopeduncular nucleus and globus pallidus in rat as demonstrated by autoradiography and horseradish peroxidase histochemistry. J Comp Neurol 177:113–123. doi:10.1002/cne.901770108
Chuhma N, Tanaka KF, Hen R, Rayport S (2011) Functional connectome of the striatal medium spiny neuron. J Neurosci 31:1183–1192. doi:10.1523/JNEUROSCI.3833-10.2011
DeFelipe J (1993) A study of NADPH diaphorase-positive axonal plexuses in the human temporal cortex. Brain Res 615:342–346. doi:10.1016/0006-8993(93)90047-Q
DeLong MR, Crutcher MD, Georgopoulos AP (1985) Primate globus pallidus and subthalamic nucleus: functional organization. J Neurophysiol 53:530–543
Deniau JM, Mailly P, Maurice N, Charpier S (2007) The pars reticulata of the substantia nigra: a window to basal ganglia output. Prog Brain Res 160:151–172
DeVito J, Anderson M (1982) An autoradiographic study of efferent connections of the globus pallidus in Macaca mulatta. Exp Brain Res 46:107–117
Dodson PD, Larvin JT, Duffell JM et al (2015) Distinct developmental origins manifest in the specialized encoding of movement by adult neurons of the external globus pallidus. Neuron 86:501–513. doi:10.1016/j.neuron.2015.03.007
Flandin P, Kimura S, Rubenstein JL (2010) The progenitor zone of the ventral medial ganglionic eminence requires Nk2–1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci 30:2812–2823. doi:10.1523/JNEUROSCI.4228-09.2010
Gerfen CR, Staines WA, Arbuthnott GW, Fibiger HC (1982) Crossed connections of the substantia nigra in the rat. J Comp Neurol 207:283–303. doi:10.1002/cne.902070308
Gittis AH, Berke JD, Bevan MD et al (2014) New roles for the external globus pallidus in basal ganglia circuits and behavior. J Neurosci 34:15178–15183. doi:10.1523/JNEUROSCI.3252-14.2014
Grace AA, Bunney BS (1983) Intracellular and extracellular electrophysiology of nigral dopaminergic neurons-1. Identification and characterization. Neuroscience 10:301–315. doi:10.1016/0306-4522(83)90135-5
Groenewegen HJ, Berendse HW, Haber SN (1993) Organization of the output of the ventral striatopallidal system in the rat: ventral pallidal efferents. Neuroscience 57:113–142. doi:10.1016/0306-4522(93)90115-V
Haber SN, Watson SJ (1985) The comparative distribution of enkephalin, dynorphin and substance P in the human globus pallidus and basal forebrain. Neuroscience 14:1011–1024. doi:10.1016/0306-4522(85)90272-6
Haber SN, Lynd-Balta E, Mitchell SJ (1993) The organization of the descending ventral pallidal projections in the monkey. J Comp Neurol 329:111–128. doi:10.1002/cne.903290108
Hernandez VM, Hegeman DJ, Cui Q et al (2015) Parvalbumin + neurons and Npas1 + neurons are distinct neuron classes in the mouse external globus pallidus. J Neurosci 35:11830–11847. doi:10.1523/JNEUROSCI.4672-14.2015
Holderith N, Lorincz A, Katona G et al (2012) Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat Neurosci 15:988–997. doi:10.1038/nn.3137
Hontanilla B, Parent A, Giménez-Amaya JM (1997) Parvalbumin and calbindin D-28k in the entopeduncular nucleus, subthalamic nucleus, and substantia nigra of the rat as revealed by double- immunohistochemical methods. Synapse 25:359–367. doi:10.1002/(SICI)1098-2396(199704)25:4<359:AID-SYN7>3.0.CO;2-9
Hoover BR, Marshall JF (2002) Further characterization of preproenkephalin mRNA-containing cells in the rodent globus pallidus. Neuroscience 111:111–125. doi:10.1016/S0306-4522(01)00565-6
Jaeger D, Gilman S, Wayne Aldridge J (1995) Neuronal activity in the striatum and pallidum of primates related to the execution of externally cued reaching movements. Brain Res 694:111–127. doi:10.1016/0006-8993(95)00780-T
Khan ZU, Gutiérrez A, Martín R, Peñafiel A, Rivera A, de la Calle A (1998) Differential regional and cellular distribution of dopamine D2-like receptors: an immunocytochemical study of subtype-specific antibodies in rat and human brain. J Comp Neurol 402:353–371. doi:10.1002/(SICI)1096-9861(19981221)402:3<353:AID-CNE5>3.0.CO;2-4
Khan ZU, Gutiérrez A, Martín R, Peñafiel A, Rivera A, de la Calle A (2000) Dopamine D5 receptors of rat and human brain. Neuroscience 100:689–699. doi:10.1016/S0306-4522(00)00274-8
Kim HF, Hikosaka O (2015) Parallel basal ganglia circuits for voluntary and automatic behaviour to reach rewards. Brain 138:1776–1800. doi:10.1093/brain/awv134
Kim R, Nakano K, Jayaraman A, Carpenter MB (1976) Projections of the globus pallidus and adjacent structures: an autoradiographic study in the monkey. J Comp Neurol 169:263–290. doi:10.1002/cne.901690302
Kim HF, Ghazizadeh A, Hikosaka O (2014) Separate groups of dopamine neurons innervate caudate head and tail encoding flexible and stable value memories. Front Neuroanat 8:120. doi:10.3389/fnana.2014.00120
Kincaid AE, Penney JB, Young AB, Newman SW (1991) Evidence for a projection from the globus pallidus to the entopeduncular nucleus in the rat. Neurosci Lett 128:121–125. doi:10.1016/0304-3940(91)90774-N
Kita H (1994) Parvalbumin-immunopositive neurons in rat globus pallidus: a light and electron microscopic study. Brain Res 657:31–41. doi:10.1016/0006-8993(94)90950-4
Kita H (2007) Globus pallidus external segment. Prog Brain Res 160:111–133
Kita H, Kita T (2001) Number, origins, and chemical types of rat pallidostriatal projection neurons. J Comp Neurol 437:438–448. doi:10.1002/cne.1294
Kita H, Kitai ST (1991) Intracellular study of rat globus pallidus neurons: membrane properties and responses to neostriatal, subthalamic and nigral stimulation. Brain Res 564:296–305. doi:10.1016/0006-8993(91)91466-E
Kita H, Kitai ST (1994) The morphology of globus pallidus projection neurons in the rat: an intracellular staining study. Brain Res 636:308–319. doi:10.1016/0006-8993(94)91030-8
Kita T, Kita H, Kitai ST (1986) Electrical membrane properties of rat substantia nigra compacta neurons in an in vitro slice preparation. Brain Res 372:21–30. doi:10.1016/0006-8993(86)91454-X
Klausberger T, Somogyi P (2008) Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321:53–57. doi:10.1126/science.1149381
Kubota Y, Karube F, Nomura M, Kawaguchi Y (2016) The diversity of cortical inhibitory synapses. Front Neural Circuits 10:1–15. doi:10.3389/fncir.2016.00027
Lammel S, Steinberg EE, Földy C et al (2015) Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron 85:429–438. doi:10.1016/j.neuron.2014.12.036
Mallet N, Micklem BR, Henny P et al (2012) Dichotomous organization of the external globus pallidus. Neuron 74:1075–1086. doi:10.1016/j.neuron.2012.04.027
Mastro KJ, Bouchard RS, Holt HAK, Gittis AH (2014) Transgenic mouse lines subdivide external segment of the globus pallidus (GPe) neurons and reveal distinct GPe output pathways. J Neurosci 34:2087–2099. doi:10.1523/JNEUROSCI.4646-13.2014
McBride RL, Larsen KD (1980) Projections of the feline globus pallidus. Brain Res 189:3–14. doi:10.1016/0006-8993(80)90003-7
Menegas W, Bergan JF, Ogawa SK et al (2015) Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife 4:1–30. doi:10.7554/eLife.2
Miyamoto Y, Fukuda T (2015) Immunohistochemical study on the neuronal diversity and three-dimensional organization of the mouse entopeduncular nucleus. Neurosci Res 94:37–49. doi:10.1016/j.neures.2015.02.006
Mounir S, Parent A (2002) The expression of neurokinin-1 receptor at striatal and pallidal levels in normal human brain. Neurosci Res 44:71–81. doi:10.1016/S0168-0102(02)00087-1
Mrzljak L, Bergson C, Pappy M et al (1996) Localization of dopamine D4 receptors in GABAergic neurons of the primate brain. Nature 381:245–248
Nambu A, Llinas R (1994) Electrophysiology of globus pallidus neurons in vitro. J Neurophysiol 72:1127–1139
Nambu A, Llinás R (1997) Morphology of globus pallidus neurons: its correlation with electrophysiology in guinea pig brain slices. J Comp Neurol 377:85–94. doi:10.1002/(SICI)1096-9861(19970106)377:1<85:AID-CNE8>3.0.CO;2-F
Nauta HJW (1979) Projections of the pallidal complex: an autoradiographic study in the cat. Neuroscience 4:1853–1873. doi:10.1016/0306-4522(79)90060-5
Nóbrega-Pereira S, Gelman D, Bartolini G et al (2010) Origin and molecular specification of globus pallidus neurons. J Neurosci 30:2824–2834. doi:10.1523/JNEUROSCI.4023-09.2010
Paladini CA, Celada P, Tepper JM (1999) Striatal, pallidal, and pars reticulata evoked inhibition of nigrostriatal dopaminergic neurons is mediated by GABA(A), receptors in vivo. Neuroscience 89:799–812. doi:10.1016/S0306-4522(98)00355-8
Parent A, Hazrati LN (1995a) Functional anatomy of the basal ganglia. I. The cortico-basal ganglia-thalamo-cortical loop. Brain Res Rev 20:91–127
Parent A, Hazrati LN (1995b) Functional anatomy of the basal ganglia. II. The place of subthalamic nucleus and external pallidium in basal ganglia circuitry. Brain Res Rev 20:128–154
Paxinos G, Watson C (2007) The rat brain in stereotaxic coordinates, 6th edn. Academic Press, San Diego
Paxinos G, Watson C, Carrive P, Kirkcaldie M, Ashwell KWS (2009) Chemoarchitectonic atlas of the rat brain, 2nd edn. Academic Press, San Diego
Petreanu L, Mao T, Sternson SM, Svoboda K (2009) The subcellular organization of neocortical excitatory connections. Nature 457:1142–1145. doi:10.1038/nature07709
Rajakumar N, Rushlow W, Naus CCG et al (1994) Neurochemical compartmentalization of the globus pallidus in the rat: an immunocytochemical study of calcium-binding proteins. J Comp Neurol 346:337–348. doi:10.1002/cne.903460303
Robledo P, Ferger J (1990) Excitatory influence of rat subthalamic nucleus to substantia nigra pars reticulata and the pallidal complex: electrophysiological data. Brain Res 518:47–54. doi:10.1016/0006-8993(90)90952-8
Sadek AR, Magill PJ, Bolam JP (2007) A single-cell analysis of intrinsic connectivity in the rat globus pallidus. J Neurosci 27:6352–6362. doi:10.1523/JNEUROSCI.0953-07.2007
Shu Y, Yu Y, Yang J, McCormick DA (2007) Selective control of cortical axonal spikes by a slowly inactivating K+ current. Proc Natl Acad Sci USA 104:11453–11458. doi:10.1073/pnas.0702041104
Smith Y, Bolam JP (1989) Neurons of the substantia nigra reticulata receive a dense GABA-containing input from the globus pallidus in the rat. Brain Res 493:160–167. doi:10.1016/0006-8993(89)91011-1
Smith AD, Bolam JP (1990a) The neural network of the basal ganglia as revealed by the study of synaptic connections of identified neurones. Trends Neurosci 13:259–265. doi:10.1016/0166-2236(90)90106-K
Smith Y, Bolam JP (1990b) The output neurones and the dopaminergic neurones of the substantia nigra receive a GABA-containing input from the globus pallidus in the rat. J Comp Neurol 296:47–64. doi:10.1002/cne.902960105
Smith Y, Bevan MD, Shink E, Bolam JP (1998) Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience 86:353–387. doi:10.1016/S0306-4522(98)00004-9
Stephenson-Jones M, Samuelsson E, Ericsson J et al (2011) Evolutionary conservation of the basal ganglia as a common vertebrate mechanism for action selection. Curr Biol 21:1081–1091. doi:10.1016/j.cub.2011.05.001
Tanahira C, Higo S, Watanabe K et al (2009) Parvalbumin neurons in the forebrain as revealed by parvalbumin-Cre transgenic mice. Neurosci Res 63:213–223. doi:10.1016/j.neures.2008.12.007
Tepper JM, Lee CR (2007) GABAergic control of substantia nigra dopaminergic neurons. Prog Brain Res 160:189–208
Tepper JM, Martin LP, Anderson DE (1995) GABAA receptor-mediated inhibition of rat substantia nigra dopaminergic neurons by pars reticulata projection neurons. J Neurosci 15:3092–3103
Watabe-Uchida M, Zhu L, Ogawa SK et al (2012) Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74:858–873. doi:10.1016/j.neuron.2012.03.017
Wheeler DW, White CM, Rees CL et al (2015) Hippocampome.org: a knowledge base of neuron types in the rodent hippocampus. eLife. doi:10.7554/eLife.09960
Wichmann T, Bergman H, DeLong MR (1994) The primate subthalamic nucleus. III. Changes in motor behavior and neuronal activity in the internal pallidum induced by subthalamic inactivation in the MPTP model of parkinsonism. J Neurophysiol 72:521–530
Acknowledgements
We thank Dr. N. Tamamaki (Kumamoto University) for providing the PV-Cre transgene construct.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Funding
This study was funded by Grants-in-Aid from The Ministry of Education, Culture, Sports, Science, and Technology (MEXT) for Scientific Research (25282247 and 15K12770 to FF; 26350983 and 16H01622 to FK; 25560435, 16H02840, 16H01623, 16K13115, 16H06543 to ST); MIC SCOPE (152107008) to ST; and for Scientific Researches on Innovative Areas “Adaptive Circuit Shift” (26112001) to FF.
Additional information
Y.-M. Oh and F. Karube equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
429_2016_1346_MOESM1_ESM.tif
Supplementary Fig. 1 Spatial distribution and co-expression of PV and Lhx6 in the GP neurons. Immunohistochemically identified PV+ (red dot), Lhx6+ (green dots), and PV+/Lhx6+ (blue dots) neurons were plotted in three sagittal planes. The magnified views of the ventral region are shown in the rightmost column. In the rightmost column, PV+/Lhx6– (red dots), PV–/Lhx6+ (green dots), and PV+/Lhx6+ (blue dots) neurons are indicated. Distribution of them was likely to be unbiased in the GP, however, Lhx6+/PV– neurons (green) were outstanding in the ventral region of the GP at LM2.9-3.9 mm (TIFF 2146 kb)
429_2016_1346_MOESM2_ESM.tif
Supplementary Fig. 2 TdTomato fluorescence, PV (blue) and Lhx6 (green) immunoreactivities in parvalbumin (PV)–Cre rats at 2 weeks after viral injection. High-magnification images displaying the viral injection site in the globus pallidus. Arrowheads indicated the Lhx6+/PV+ neurons. Note that Lhx6+/PV– neurons (arrows) were rarely infected. Photos were taken with a confocal microscope (FV-1200, Olympus, Tokyo, Japan) under the conditions described in the Materials and Methods. Scale bars: 20 µm (TIFF 10991 kb)
429_2016_1346_MOESM3_ESM.tif
Supplementary Fig. 3 AAV vector used here did not label neurons retrogradely. Fluorescent images of the SN (A), EP (B), and striatum (C) of a rat after AAV injection into the GP. TdTomato labelled axons issuing from PV-GP neurons were observed in all images. Importantly, no retrograde labeling of PV neurons was observed. A neuron-like shape of fluorescence was investigated at some locations in SNr (A2) and EP (B), however, magnified views combined with PV immunoreactivity (green) revealed that these pseudo-cell like fluorescence was not AAV-infected neurons. Actually they were unlabeled neurons which were surrounded by dense PV-GP axons. (C) In striatum, no such image was observed due to relatively weak PV-GP projections. Photos were taken with a confocal microscope (FV-1200, Olympus, Tokyo, Japan) under the conditions described in the Materials and Methods (TIFF 30222 kb)
429_2016_1346_MOESM4_ESM.tif
Supplementary Fig. 4 Fluorescent images of axons from parvalbumin (PV)–globus pallidus (GP) neurons in the substantia nigra pars compacta (SNc) and reticulata (SNr). (A) Overlay (the most right) of PV-GP axons (red), tyrosine hydroxylase (TH, blue) and PV (green) immunofluorescence in case b. Using TH and PV immunoreactivity SNc and SNr were identified. (B) Fluorescent images of axons from PV-GP neurons in the SN from 5 PV-Cre rats (cases a–e). Fluorescent images in the left column display the virus injection sites in the GP. Five latero-medial planes of SN images are arranged from left (the most lateral plane) to right (the most medial plane), ranged from 1.55 to 2.9 mm. Note that dense boutons of PV-GP axons emerged in the border region between the SNc and SNr. See also Fig. 5 and 6. Photos were taken with an epifluorescent microscope (BX-61, Olympus, Tokyo, Japan) under the conditions described in the Materials and Methods. Scale bars: 500 µm in GP and 200 µm in SN (TIFF 48117 kb)
429_2016_1346_MOESM5_ESM.tif
Supplementary Fig. 5 Procedure for subdivision of the SN area. (A) The area of SNc was defined by TH immunoreactivity. (B) The leftmost and rightmost positions of the polygon were identified, respectively (green dots). (C) The bottom polygonal line (red) was defined as the border between SNc and SNr. (D) A line connecting coordinates of the horizontal axis of the positions was equally divided into 100 positions (top dots). (E) At each divided position, points that bisect a perpendicular line intersecting the polygon were identified (ex. blue dot). A polygonal line that connects all the bisection points was defined as a border between area I and area II (green polygonal line). (F) Points that are arranged symmetrically against the bisection point in the vertical direction with the border between SNc and SNr as the axis of symmetry at each divided location were identified, and then a polygonal line that connects the points was defined as a border between area III and area IV (blue polygonal line). (G) Points that are arranged symmetrically against the point on the border between SNc and SNr in the vertical direction with the border between area III and area IV as the axis of symmetry at each divided location were identified, and then a polygonal line that connects the points was defined as a bottom edge of all the area (blue dotted polygonal line). (H) The identified four areas (I – IV) of SN (TIFF 1795 kb)
429_2016_1346_MOESM6_ESM.tif
Supplementary Fig. 6 Fluorescent images of CB, CCK, CR or NOS-positive cell body and axons from PV-GP neurons in the SN. Overlay of PV-GP axons (red), CB, CCK, CR or NOS (green) immunofluorescence in SN (A), SNc (B) and SNr (C). Note that PV-GP axons formed basket-like appositions with CCK (b) and CR (c) but CB (a) immunoreactive somata in the SNc and with NOS (C) immunoreactive soma in the SNr. PV-GP axons in the border region between the SNc and SNr formed basket-like appositions with CCK and CR somata. Photos were taken with a confocal microscope (FV-1200, Olympus, Tokyo, Japan) under the conditions described in the Materials and Methods. Scale bar: low magnification, 200 µm; high magnification, 20 µm (TIFF 33669 kb)
429_2016_1346_MOESM7_ESM.tif
Supplementary Fig. 7 Estimated distances between the centers of the TH-positive cell bodies and PV-GP boutons. Sample IDs are indicated in the topmost section, which are the same as those in Fig. 6. (A) Average frequency of the number of boutons per TH-positive cell as a function of the normalized distance between the cell center and edge. Red bars indicate standard errors of the mean. (B) Bar graph shows the sum total of boutons for each cell within the near-center and near-edge regions, respectively. The borderline between the cell center and edge is set at the midpoint. Red bars indicate standard deviations. Wilcoxon rank-sum test, *: p<0.05, ***: p<0.001 (TIFF 2453 kb)
429_2016_1346_MOESM8_ESM.tif
Supplementary Fig. 8 Estimated distances between the centers of the PV-positive cell bodies and PV-GP boutons. Sample IDs are indicated in the topmost section, which are the same as those in Fig. 6. (A) Average frequency of the number of boutons per PV-positive cell as a function of the normalized distance between the cell center and edge. Red bars indicate standard errors of the mean. (B) Bar graph shows the sum total of boutons for each cell within the near-center and near-edge regions, respectively. The borderline between the cell center and edge is set at the midpoint. Red bars indicate standard deviations. Wilcoxon rank-sum test, *: p<0.05, **: p<0.01, ***: p<0.001 (TIFF 1523 kb)
429_2016_1346_MOESM9_ESM.tif
Supplementary Fig. 9 Estimated distances between the centers of the CB, CCK, CR or NOS-positive cell bodies and PV-GP boutons. Sample IDs are indicated in the topmost section, which are the same as those in Fig. 6. (A) Average frequency of the number of boutons per CB (f), CCK (g), CR (h) or NOS (i) -positive cell as a function of the normalized distance between the cell center and edge. Red bars indicate standard errors of the mean. (B) Bar graph shows the sum total of boutons for each cell within the near-center and near-edge regions, respectively. The borderline between the cell center and edge is set at the midpoint. Red bars indicate standard deviations. Wilcoxon rank-sum test, ***: p<0.001. Since for CB and NOS - positive cells, few boutons were located around the cell body, statistical test could not be performed (TIFF 1433 kb)
Rights and permissions
About this article
Cite this article
Oh, YM., Karube, F., Takahashi, S. et al. Using a novel PV-Cre rat model to characterize pallidonigral cells and their terminations. Brain Struct Funct 222, 2359–2378 (2017). https://doi.org/10.1007/s00429-016-1346-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-016-1346-2