Abstract
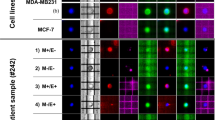
Breast cancer epithelial cells with the CD44+/CD24−/low phenotype possess tumor-initiating cells and epithelial-mesenchymal transition (EMT) capacity. Massive parallel sequencing can be an interesting approach to deepen the molecular characterization of these cells. We characterized CD44+/CD24−/cytokeratin(Ck)+/CD45− cells isolated through flow cytometry from 43 biopsy and 6 mastectomy samples harboring different benign and malignant breast lesions. The Ion Torrent Ampliseq Cancer Hotspot panel v2 (CHPv2) was used for the identification of somatic mutations in the DNA extracted from isolated CD44+/CD24−/Ck+/CD45− cells. E-Cadherin and vimentin immunohistochemistry was performed on sections from the corresponding formalin-fixed, paraffin-embedded (FFPE) blocks. The percentage of CD44+/CD24−/Ck+/CD45− cells increased significantly from non-malignant to malignant lesions and in association with a significant increase in the expression of vimentin. Non-malignant lesions harbored only a single-nucleotide polymorphism (SNP). Mutations in the tumor suppressor p53 (TP53), NOTCH homolog 1 (NOTCH1), phosphatase and tensin homolog (PTEN), and v-akt murine thymoma viral oncogene homolog 1 (AKT1) genes were found in isolated CD44+/CD24−/Ck+/CD45− cells from ductal carcinomas in situ (DCIS). Additional mutations in the colony-stimulating factor 1 receptor (CSF1R), ret proto-oncogene (RET), and TP53 genes were also identified in invasive ductal carcinomas (IDCs). The use of massive parallel sequencing technology for this type of application revealed to be extremely effective even when using small amounts of DNA extracted from a low number of cells. Additional studies are now required using larger cohorts to design an appropriate mutational profile for this phenotype.



Similar content being viewed by others
References
Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H (2005) Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res 11:1154–1159
Ginestier C, Min MH, Charafe-Jauffret E et al (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1:555–567
Symmans WF, Liu J, Knowles DM, Inghirami G (1995) Breast cancer heterogeneity: evaluation of clonality in primary and metastatic lesions. Hum Pathol 26:210–216
Martelotto LG, Ng CK, Piscuoglio S, Weigelt B, Reis-Filho JS (2014) Breast cancer intra-tumor heterogeneity. Breast Cancer Res Treat 16:210
Neumeister v, Agarwal S, Bordeaux J, Camp RL, Rimm DL (2010) In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol 176:2131–2138
Xiao Y, Ye Y, Yearsley K, Jones S, Barsky SH (2008) The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Am J Pathol 173:561–574
Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL (2009) High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med 13:2236–2252
Da Cruz PA, Marques O, Rosa AM, Faria MDF, Rema A, Lopes C (2014) Co-expression of stem cell markers ALDH1 and CD44 in non-malignant and neoplastic lesions of the breast. Anticancer Res 34:1427–1434
Da Cruz PA, Marques O et al (2016) Characterization of CD44+ ALDH1+ Ki-67− cells in non-malignant and neoplastic lesions of the breast. Anticancer Res 36:4629–4638
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100:3983–3988
Ponti D, Costa A et al (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 65:5506–5511
Fillmore CM, Kuperwasser C (2008) Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 10:R25
Sheridan C, Kishimoto H et al (2006) CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 8:R59
Balic M et al (2006) Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res 12:5615–5621
Theodoropoulos PA, Polioudaki H et al (2010) Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett 288:99–106
Wang N, Shi L et al (2012) Detection of circulating tumor cells and tumor stem cells in patients with breast cancer by using flow cytometry. Tumor Biol 33:561–569
Hernandez L, Wilkerson PM, Lambros MB et al (2012) Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol 227:42–52
Navin N, Kendall J, Troge J et al (2011) Tumour evolution inferred by single-cell sequencing. Nature 472:90–94
Jorge RF (2009) Next-generation sequencing. Breast Cancer Res Treat 11:S12
Liu X, Mody K, de Abreu FB et al (2014) Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin Chem 60:1004–1011
Amato E, dal Molin M, Mafficini A et al (2014) Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol 233:217–227
Cheang MCU, Chia SK, Voduc D et al (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101:736–750
Riethdorf S, Fritsche H et al (2007) Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the Cell Search system. Clin Cancer Res 13:920–928
Cristofanilli M, Budd GT et al (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351:781–791
Hayes DF, Cristofanilli M et al (2006) Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res 15:4218–4224
Cohen SJ, Punt CJ et al (2008) Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 26:3213–3221
De Bono JS, Scher et al (2008) Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 14:6302–6309
Hardt O, Wild S et al (2012) Highly sensitive profiling of CD44+/CD24− breast cancer stem cells by combining global mRNA amplification and next generation sequencing: evidence for a hyperactive PI3K pathway. Cancer Lett 325:165–174
Geens M, Van de Velde H et al (2007) The efficiency of magnetic-activated cell sorting and fluorescence-activated cell sorting in the decontamination of testicular cell suspensions in cancer patients. Hum Reprod 22:733–742
Bane A, Viloria-Petit A, Pinnaduwage D, Mulligan AM, O’Malley FP, Andrulis IL (2013) Clinical–pathologic significance of cancer stem cell marker expression in familial breast cancers. Breast Cancer Res Treat 140:195–205
Makki J, Myint O, Wynn AA, Samsudin AT, John DV (2014) Expression distribution of cancer stem cells, epithelial to mesenchymal transition, and telomerase activity in breast cancer and their association with clinicopathologic characteristics. Clin Med Insights Pathol 8:1–16
Vuoriluoto K, Haugen H, Kiviluoto S et al (2011) Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 30:1436–1448
Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz T, Rosen JM (2007) WNT/β-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A 104:618–623
Choi Y, Lee HJ, Jang MH et al (2013) Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum Pathol 44:2581–2589
Diehn M, Cho RW, Lobo NA et al (2009) Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458:780–783
de Leng W, Gadellaa-van Hooijdonk CG, Barendregt-Smouter FA, Koudijs MJ et al (2016) Targeted next generation sequencing as a reliable diagnostic assay for the detection of somatic mutations in tumours using minimal DNA amounts from formalin fixed paraffin embedded material. PLoS One 11:e0149405
Chang CJ, Chao CH, Xia W et al (2011) p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 13:317–323
Farnie G, Clarke RB, Spence K et al (2007) Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst 99:616–627
Koh M, Woo Y, Valiathan RR et al (2015) Discoidin domain receptor 1 is a novel transcriptional target of ZEB1 in breast epithelial cells undergoing H-Ras-induced epithelial to mesenchymal transition. Intl J Cancer 6:E508–E520
Vuoriluoto K, Haugen H, Kiviluoto S et al (2011) Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 12:1436–1448
Zhou J, Wulfkuhle J, Zhang H et al (2007) Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci 104:16158–16163
Zhu QS, Rosenblatt K, Huang KL et al (2011) Vimentin is a novel AKT1 target mediating motility and invasion. Oncogene 30:457–470
Tsongalis GJ, Peterson JD et al (2014) Routine use of the Ion Torrent AmpliSeq™ Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clinical Chem Lab Med 52:707–714
Kacinski BM, Scata KA, Carters D et al (1991) FMS (CSF-l receptor) and CSF-l transcripts and protein are expiessed. Oncogene 6:941–952
Sapi E, Flick MB, Rodov S et al (1996) Independent regulation of invasion and anchorage-independent growth by different autophosphorylation sites of the macrophage colony-stimulating factor 1 receptor. Cancer Res 56:5704–5712
Gao J, Aksoy BA, Dresdner G et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 269:pl1
Gattelli A, Nalvarte I, Boulay A et al (2013) Ret inhibition decreases growth and metastatic potential of estrogen receptor positive breast cancer cells. EMBO Mol Med 5:1335–1350
Kan Z, Jaiswal BS, Stinson J et al (2010) Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466:869–873
Mimori K, Inoue H, Shiraishi T et al (2002) A single-nucleotide polymorphism of SMARCB1 in human breast cancers. Genomics 80:254–258
Acknowledgments
This study was funded by the Foundation for Science and Technology (Portugal). Scholarships references: SFRH/BD/74307/2010 (ACP) and SFRH/BD/2011/78184 (OM). The authors would like to thank the invaluable help of Manuela Certo and Maria José Oliveira during breast tissue collection.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This project was approved by the following ethical boards: Porto Hospital Centre Research Ethics Health Committee (reference 203-CES) and by Porto Hospital Centre Department of Education, Development and Research (reference 135-DEFI).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Margarida Lima and Carlos Lopes contributed equally to the supervision of this work.
Rights and permissions
About this article
Cite this article
Da Cruz Paula, A., Leitão, C., Marques, O. et al. Molecular characterization of CD44+/CD24−/Ck+/CD45− cells in benign and malignant breast lesions. Virchows Arch 470, 311–322 (2017). https://doi.org/10.1007/s00428-017-2068-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-017-2068-4