Abstract
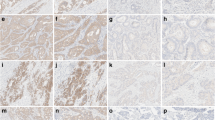
The activating BRAF V600 mutation is a well-established negative prognostic biomarker in metastatic colorectal carcinoma (CRC). A recently developed monoclonal mouse antibody (clone VE1) has been shown to detect reliably BRAF V600E mutated protein by immunohistochemistry (IHC). In this study, we aimed to compare the detection of BRAF V600E mutations by IHC, Sanger sequencing (SaS), and ultra-deep sequencing (UDS) in CRC. VE1-IHC was established in a cohort of 68 KRAS wild-type CRCs. The VE1-IHC was only positive in the three patients with a known BRAF V600E mutation as assessed by SaS and UDS. The test cohort consisted of 265 non-selected, consecutive CRC samples. Thirty-nine out of 265 cases (14.7 %) were positive by VE1-IHC. SaS of 20 randomly selected IHC negative tumors showed BRAF wild-type (20/20). Twenty-four IHC-positive cases were confirmed by SaS (24/39; 61.5 %) and 15 IHC-positive cases (15/39; 38.5 %) showed a BRAF wild-type by SaS. UDS detected a BRAF V600E mutation in 13 of these 15 discordant cases. In one tumor, the mutation frequency was below our threshold for UDS positivity, while in another case, UDS could not be performed due to low DNA amount. Statistical analysis showed sensitivities of 100 % and 63 % and specificities of 95 and 100 % for VE1-IHC and SaS, respectively, compared to combined results of SaS and UDS. Our data suggests that there is high concordance between UDS and IHC using the anti-BRAFV600E (VE1) antibody. Thus, VE1 immunohistochemistry is a highly sensitive and specific method in detecting BRAF V600E mutations in colorectal carcinoma.





Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA: A Cancer Journal for Clinicians 61(2):69–90. doi:10.3322/caac.20107
Ries L, Young J, Keel G, Eisner M, Lin Y, Horner M-J (2007) SEER survival monograph: cancer survival among adults: U.S. SEER Program, 1988–2001, Patient and Tumor Characteristics. National Cancer Institute, SEER Program, Bethesda, MD
Van Cutsem E, Kohne CH, Lang I et al (2011) Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 29(15):2011–2019. doi:10.1200/JCO.2010.33.5091
Di Nicolantonio F, Martini M, Molinari F et al (2008) Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 26(35):5705–5712. doi:10.1200/JCO.2008.18.0786
Bokemeyer C, Cutsem EV, Rougier P et al (2012) Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer 48(10):1466–1475. doi:10.1016/j.ejca.2012.02.057
Safaee Ardekani G, Jafarnejad SM, Tan L, Saeedi A, Li G (2012) The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis. PLoS ONE 7(10):e47054. doi:10.1371/journal.pone.0047054
Roth AD, Tejpar S, Delorenzi M et al (2010) Prognostic role of KRAS and BRAF in Stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. Journal of Clinical Oncology 28(3):466–474. doi:10.1200/jco.2009.23.3452
Fransén K, Klintenäs M, Österström A, Dimberg J, Monstein H-J, Söderkvist P (2004) Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis 25(4):527–533. doi:10.1093/carcin/bgh049
Davies H, Bignell GR, Cox C et al (2002) Mutations of the BRAF gene in human cancer. Nature 417(6892):949–954. doi:10.1038/nature00766
Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE (2002) Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418(6901):934–934. doi:10.1038/418934a
Behl AS, Goddard KA, Flottemesch TJ, Veenstra D, Meenan RT, Lin JS, Maciosek MV (2012) Cost-Effectiveness Analysis of Screening for KRAS and BRAF Mutations in Metastatic Colorectal Cancer. J Natl Cancer Inst 104(23):1785–1795. doi:10.1093/jnci/djs433
Blank PR, Moch H, Szucs TD, Schwenkglenks M (2011) KRAS and BRAF mutation analysis in metastatic colorectal cancer: a cost-effectiveness analysis from a Swiss perspective. Clinical Cancer Research 17(19):6338–6346. doi:10.1158/1078-0432.CCR-10-2267
Lamy A, Blanchard F, Le Pessot F et al (2011) Metastatic colorectal cancer KRAS genotyping in routine practice: results and pitfalls. Mod Pathol 24(8):1090–1100. doi:10.1038/modpathol.2011.60
Anderson S, Bloom KJ, Vallera DU et al (2012) Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Archives of Pathology & Laboratory Medicine 136(11):1385–1391. doi:10.5858/arpa.2011-0505-OA
Rechsteiner M, von Teichman A, Rüschoff JH et al (2013) KRAS, BRAF, and TP53 deep sequencing for colorectal carcinoma patient diagnostics. The Journal of Molecular Diagnostics 15(3):299–311. doi:10.1016/j.jmoldx.2013.02.001
Raab SS (2000) The cost-effectiveness of immunohistochemistry. Archives of Pathology & Laboratory Medicine 124(8):1185–1191. doi:10.1043/0003-9985(2000)124<1185:tceoi>2.0.co;2
Capper D, Preusser M, Habel A et al (2011) Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol 122(1):11–19. doi:10.1007/s00401-011-0841-z
Andrulis M, Penzel R, Weichert W, von Deimling A, Capper D (2012) Application of a BRAF V600E mutation-specific antibody for the diagnosis of hairy cell leukemia. The American Journal of Surgical Pathology 36(12):1796–1800. doi:10.1097/PAS.0b013e3182549b50
Bösmüller H, Fischer A, Pham DL et al (2013) Detection of the BRAFV600E mutation in serous ovarian tumors: a comparative analysis of immunohistochemistry with a mutation-specific monoclonal antibody and allele-specific PCR. Human Pathology 44(3):329–335. doi:10.1016/j.humpath.2012.07.010
Bullock M, O'Neill C, Chou A et al (2012) Utilization of a MAB for BRAFV600E detection in papillary thyroid carcinoma. Endocrine-Related Cancer 19(6):779–784. doi:10.1530/erc-12-0239
Capper D, Berghoff A, Magerle M et al (2012) Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol 123(2):223–233. doi:10.1007/s00401-011-0887-y
Ilie M, Long E, Hofman V et al (2012) Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Annals of Oncology 24(3):742–748. doi:10.1093/annonc/mds534
Koperek O, Kornauth C, Capper D et al (2012) Immunohistochemical detection of the BRAF V600E-mutated protein in papillary thyroid carcinoma. The American Journal of Surgical Pathology 36(6):844–850. doi:10.1097/PAS.0b013e318246b527
Long GV, Wilmott JS, Capper D et al (2013) Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. The American Journal of Surgical Pathology 37(1):61–65. doi:10.1097/PAS.0b013e31826485c0
Preusser M, Capper D, Berghoff AS et al (2013) Expression of BRAF V600E mutant protein in epithelial ovarian tumors. Applied immunohistochemistry & molecular morphology 21(2):159–164
Sahm F, Capper D, Preusser M et al (2012) BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood 120(12):e28–e34. doi:10.1182/blood-2012-06-429597
Kononen J, Bubendorf L, Kallioniemi A et al (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4(7):844–847. doi:10.1038/nm0798-844
Colomba E, Hélias-Rodzewicz Z, Von Deimling A et al (2013) Detection of BRAF p.V600E Mutations in Melanomas: Comparison of Four Methods Argues for Sequential Use of Immunohistochemistry and Pyrosequencing. The Journal of Molecular Diagnostics 15(1):94–100. doi:10.1016/j.jmoldx.2012.09.001
Affolter K, Samowitz W, Tripp S, Bronner MP (2013) BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes, Chromosomes and Cancer 52(8):748–752. doi:10.1002/gcc.22070
Adackapara CA, Sholl LM, Barletta JA, Hornick JL (2013) Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology 63(2):187–193. doi:10.1111/his.12154
De Roock W, Claes B, Bernasconi D et al (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The Lancet Oncology 11(8):753–762. doi:10.1016/S1470-2045(10)70130-3
Capper D, Voigt A, Bozukova G et al (2013) BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer 133(7):1624–1630. doi:10.1002/ijc.28183
Baldus SE, Schaefer K-L, Engers R, Hartleb D, Stoecklein NH, Gabbert HE (2010) Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clinical Cancer Research 16(3):790–799. doi:10.1158/1078-0432.ccr-09-2446
Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, Quirke P (2011) Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Analytical Cellular Pathology 34(1):61–66. doi:10.3233/acp-2011-0005
Guerra A, Fugazzola L, Marotta V et al (2012) A high percentage of BRAF V600E alleles in papillary thyroid carcinoma predicts a poorer outcome. Journal of Clinical Endocrinology & Metabolism 97(7):2333–2340. doi:10.1210/jc.2011-3106
Sinicrope FA, Smyrk TC, Tougeron D et al (2013) Mutation-specific antibody detects mutant BRAFV600E protein expression in human colon carcinomas. Cancer 119(15):2765–2770. doi:10.1002/cncr.28133
Acknowledgements
We thank Dr. Adriana von Teichman, Marion Bawohl, and Sonja Brun-Schmid for their excellent technical assistance. We are very grateful to Prof. Dieter Zimmermann for his critical proof-reading of the manuscript.
Conflict of interests
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Matthias Rössle and Michèle Sigg contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1211 kb)
Rights and permissions
About this article
Cite this article
Rössle, M., Sigg, M., Rüschoff, J.H. et al. Ultra-deep sequencing confirms immunohistochemistry as a highly sensitive and specific method for detecting BRAF V600E mutations in colorectal carcinoma. Virchows Arch 463, 623–631 (2013). https://doi.org/10.1007/s00428-013-1492-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-013-1492-3