Abstract
KRAS mutation testing is mandatory before prescribing anti-epidermal growth factor monoclonal antibodies in the treatment of advanced colorectal cancer. We describe the performance of a TaqMelt polymerase chain reaction (PCR) assay—the cobas® KRAS Mutation Test—designed to detect 19 mutations in codons 12, 13, and 61. The limit of detection was determined using DNA blends from cell lines, plasmids, and formalin-fixed paraffin-embedded tissue specimens. Assay performance was compared to Sanger sequencing using a panel of 188 specimens. Discordant specimens were subjected to next generation pyrosequencing (454). Assay repeatability was assessed using a panel of six specimens. A >95% correct mutation call rate was obtained in all specimen types with ~5% mutant alleles at DNA inputs of 0.8–6.3 ng per PCR reaction; 100% detection rate was observed at the recommended DNA input of 50 ng. The positive percent agreement with Sanger was 97.5% (79/81) for codons 12/13 and 85.7% (6/7) for codon 61. Negative percent agreement was 94.4% (101/107) for codon 12/13 and 99.4% (180/181) for codon 61. Nine of 10 discordant specimens yielded 454 results consistent with the cobas® results. With repeated testing, the assay showed a correct call rate of 100% (192/192) for all operators, instruments, reagent lots, and days tested. The cobas® test detects KRAS mutations in codons 12, 13, and 61 at a limit of detection of <5%. The PCR assay was more sensitive and specific than Sanger sequencing, and performance was highly reproducible. Test performance was not influenced by various endogenous interfering substances or common gut microbes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Mutations in the KRAS gene that lead to its constitutive activation have been identified in 24–43% of colorectal cancer (CRC) tumors and are common in other tumor types such as pancreatic, lung, thyroid, and myeloid leukemia [1–3]. These gain of function mutations lead to epidermal growth factor receptor (EGFR)-independent activation of intracellular signaling pathways, resulting in tumor cell proliferation, protection against apoptosis, increased invasion and metastasis, and activation of tumor-induced angiogenesis [4]. The presence of activating mutations has been shown in randomized clinical trials to play a crucial role in predicting non-responsiveness in patients with advanced CRC who receive the anti-EGFR monoclonal antibodies cetuximab and panitumumab [5–9]. The majority of activating mutations in CRC tumors occur in codons 12 (~82%) and 13 (~17%) of exon 2 of the KRAS gene; however, mutations in codon 61 (exon 3) have also been described [10]. Codon 61 mutations have been shown to result in constitutive activation of KRAS just as codon 12 and 13 mutations do [11, 12], and limited clinical data indicate that codon 61 mutations may predict non-response to anti-EGFR monoclonal antibody therapy [13–15].
The high level evidence supporting the clinical utility of KRAS mutation status in predicting response to anti-EGFR monoclonal antibodies has led to specific requirements for KRAS mutation testing by European and US regulatory authorities [16, 17]. Major oncology guidelines, including those of the European Society for Medical Oncology [18] and the American Society of Clinical Oncology [19], have all embraced the recommendation that these antibodies be restricted to patients with KRAS wild-type colorectal cancers.
Bi-directional Sanger sequencing, which theoretically can identify all possible mutations in an exon, has been a common reference method for detecting somatic mutations in tumor specimens. However, Sanger sequencing suffers from limited sensitivity for low-level mutant alleles, particularly in formalin-fixed paraffin-embedded tissue (FFPET) specimens, and has a slow turnaround time [20]. A number of other KRAS mutation detection methods, including allele-specific PCR assays, high-resolution melting analysis, and pyrosequencing, have been developed [21, 22]. These other methods have been reported to have varying levels of sensitivity and specificity for detecting KRAS mutations and there is currently no uniform standard for KRAS mutation testing.
There is a clear need for a well-validated, robust, and reproducible method. The European Union in vitro Diagnostic (IVD) Medical Device Directive 98/79/EC requires that IVDs “achieve the performance…stated by the manufacturer” for a variety of test attributes such as limits of detection, analytical sensitivity and specificity, reproducibility/repeatability, and potential interfering and cross-reacting substances. Aligning with the European Union IVD directives and optimizing the analytical performance of a mutation assay requires thorough validation testing on different specimen types (including the intended use of FFPET specimens) as well as a comprehensive understanding of the impact of specimen attributes, such as the tumor content, the levels of amplifiable DNA, necrosis, and the influence of endogenous and exogenous interfering substances.
We set out to design a robust, accurate molecular diagnostic test to identify patients who have CRC tumors with somatic mutations in codons 12 and 13 of exon 2 or 61 of exon 3 of the KRAS gene and who are therefore unlikely to benefit from therapy with anti-EGFR monoclonal antibodies. The cobas® KRAS Mutation Test (Roche Molecular Systems, Branchburg, NJ, USA), for use with the cobas® 4800 System v2.0 is a TaqMelt™ real-time PCR test intended to detect 19 mutations in codons 12 and 13 of exon 2 and 61 of exon 3 of the KRAS gene using DNA derived from FFPET from human CRC tumors. In this report, we present data on the analytic performance of the cobas® test.
Material and methods
Materials
FFPET specimens from CRC tumors were purchased from US commercial vendors: Discovery Life Sciences, Inc. (Los Osos, CA, USA); BioServe (Beltsville, MD, USA); ProteoGenex (Culver City, CA, USA); CureLine, Inc. (San Francisco, CA, USA); and Indivumed, Inc. (Kensington, MD, USA). The FFPET specimens from vendors were from CRC stages I to IV with a majority from stages II and III. Estimated tumor content ranged from 25% to 80%.
Cobas® KRAS mutation test
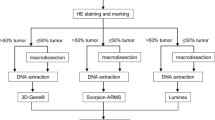
The cobas® KRAS Mutation Test kit (cobas® test) is a TaqMelt™-based PCR assay (Fig. 1). The testing process consists of two main steps: manual specimen preparation to obtain genomic DNA from FFPET and PCR amplification and detection of target DNA using complementary primer pairs and two oligonucleotide probes labeled with different fluorescent dyes. The PCR primers are designed to amplify defined sections of exons 2 and 3 of the KRAS gene, and fluorescently labeled oligonucleotide probes detect mutations in codons 12, 13, and 61, which fall within these exons.
Assay workflow. Two 5-μm sections are prepared from each FFPET specimen—one section is used for H&E staining to assess tumor content and the other section is used for DNA isolation. If the tumor content is greater than 10%, which all of the specimens for the studies in this report were, then no macro-dissection is required. Genomic DNA is isolated using the cobas® DNA Sample Preparation Kit and two 25-μL aliquots (one aliquot for codon 12/13 of exon 2 and the other aliquot for codon 61 of exon 3) of the resulting extract that is adjusted to 2 ng/μL of DNA are amplified using cobas z 480 analyzer. Change in fluorescence during melting steps is automatically analyzed for melting temperatures and peak heights for mutants and wild types to determine mutation status of each specimen
DNA extraction: DNA is extracted from a single, deparaffinized 5 μm section of FFPET following the standard procedure described in the cobas® DNA Sample Preparation Kit package insert. A deparaffinized specimen is lysed by incubation at an elevated temperature with a protease and chaotropic lysis/binding buffer that releases nucleic acid and protects the released genomic DNA from DNases. Subsequently, isopropanol is added to the lysis mixture that is then centrifuged through a column with a glass fiber filter. Unbound substances, such as salts, proteins, and other cellular impurities, are removed by centrifugation. The adsorbed nucleic acids are washed and then eluted with an aqueous solution. The amount of genomic DNA is spectrometrically determined and adjusted to a fixed concentration of 2 ng/μL.
PCR amplification and detection: The target DNA is amplified and detected on the cobas® 4800 system, which measures the fluorescence generated by specific PCR products, using the amplification and detection reagents provided in the cobas® KRAS Mutation Test kit (Fig. 1). Mutation detection is achieved by melting curve analysis, using an automated result interpretation software package (KRAS Assay Specific Analysis Package v1.0). Based on melting temperatures and peak heights for mutants and wild types, mutation status of each specimen was determined. A mutant control, a negative control, and a calibrator are included in each run to confirm the validity of the run. Amplification, detection, quality control analysis, and result interpretation are automated on the system; raw data and melting curves are not reported. The DNA isolation, amplification/detection, and result reporting can be performed in less than 8 h. The test kit is configured such that users running a minimum of three specimens can achieve the full number of 24 reportable results per kit; as many as 45 specimens can be processed in a single test run.
Sanger sequencing
DNA prepared from FFPET specimens using the same extraction method as described for the cobas® test were sequenced by 2X bi-directional Sanger sequencing by a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (SeqWright, Houston, TX) using a validated protocol.
454 quantitative massively parallel pyrosequencing
Specimens which gave discordant cobas® test and Sanger test results were retested using a quantitative massively parallel pyrosequencing method (454 GS Titanium, 454 Life Sciences, Branford, CT) by a CLIA-certified laboratory (SeqWright, Houston, TX) using a validated protocol. Additionally, a randomly selected subset of specimens with concordant cobas® test and Sanger results were tested by 454 sequencing to confirm the accuracy of mutation status. The validated limit of detection for the 454 KRAS protocol was 1%.
Limit of detection
The limit of detection (LOD) of the cobas® test was evaluated for one predominant mutation in codon 12 (GAT), codon 13 (GAC), and codon 61 (CAC) of the KRAS gene with ~5% mutation. Testing on 24–72 replicates per panel member at different levels of DNA input was conducted using the following specimen types:
-
FFPET specimens,
-
DNA extracted from CRC FFPET mutant and wild-type specimens,
-
DNA extracted and blended from two CRC mutant and wild-type cell lines for codons 12 and 13, and
-
DNA from one mutant plasmid for codon 61 blended with wild-type KRAS cell line DNA.
Testing was performed using three lots of the cobas® test kit and one lot of the cobas® DNA Sample Preparation Kit. DNA extracted from each specimen type was then serially diluted to DNA input levels ranging from 50.0 to 0.4 ng per reaction well, in order to simulate varying degrees of DNA degradation that may occur with formalin fixation. 454 sequencing was used to assess percent mutant alleles for DNA extracted from each specimen type. Sensitivity was determined based on a correct mutation call rate of ≥95% and by Probit analysis.
In a separate analysis, the LOD was evaluated for 16 additional KRAS mutation genotypes. Due to the lack of naturally occurring specimens with ~5% mutant alleles, FFPET DNA and plasmid blends at ~5% mutation (as assessed by 454 sequencing) were prepared including:
-
Five FFPET blends representing five KRAS codon 12 mutations,
-
Five blends (four plasmid and one FFPET) representing five KRAS codon 13 mutations, and
-
Six blends (two plasmid and four FFPET) representing six KRAS codon 61 mutations.
Before cobas® testing, the mutation status and mutation percentage of the specimens were verified by 2X bi-directional Sanger sequencing and 454 sequencing, respectively.
Analytic sensitivity and specificity—methods correlation with Sanger sequencing
The analytic performance of the cobas® test was evaluated by comparing two lots of the cobas® KRAS test to 2X bi-directional Sanger Sequencing using 188 vendor-purchased CRC FFPET specimens. Each cobas® test reagent lot was paired with a separate lot of the cobas® DNA Sample Preparation Kit. Specimens were tested in a blinded fashion for the presence of codon 12, 13, and 61 mutations. Positive and negative percent agreements between the cobas® test and Sanger test results were determined. Discrepant analysis was performed by testing specimens with discordant cobas® test and Sanger results with 454 sequencing for resolution. In addition, 50 randomly selected specimens with concordant Sanger and cobas® test results were tested by 454 sequencing in order to provide further verification of the correct call rate.
Repeatability
The repeatability of the cobas® test was evaluated across different reagent lots, days, operators, and instruments with six FFPET CRC specimens, two KRAS wild-type, and four KRAS mutation positive. The materials used in the repeatability testing are:
-
One cobas® DNA Sample Preparation Kit lot,
-
Two cobas® KRAS Mutation Test Kit lots,
-
Two operators, and
-
Four cobas® 4800 System v.2.0.
Each operator performed one run per reagent lot per day for 4 days for a total of 16 runs with two replicates of each of the six specimens.
Interfering endogenous substances
The effects of endogenous substances, hemoglobin and triglycerides, on the performance of the cobas® test were investigated in 10 CRC FFPET specimens. The specimens were selected for mutation status based on Sanger and/or 454 sequencing. Five specimens were KRAS mutation positive and five were wild type. Four 5-μm sections were extracted for each specimen, one each in the following:
-
Tissue Lysis Buffer + Proteinase K (TLB/PK) (control),
-
TLB/PK + saline (solvent control),
-
TLB/PK + 2 g/L hemoglobin (CLSI-recommended test concentrations), and
-
TLB/PK + 37 mM triglycerides.
Hemoglobin and triglycerides were added during the lysis step of DNA extraction and were not subjected to deparaffinization. All DNA extracts were tested for interference of mutation detection using the cobas® test.
Interference by common colon-related microorganisms
The effects of colon-related microorganisms on the performance of the cobas® test were investigated in 10 FFPET CRC specimens. The specimens were selected for mutation status based on Sanger and/or 454 sequencing. Five specimens were KRAS mutation positive and five were wild type. Approximately one million colony-forming units of Bacteroides caccae, Prevotella intermedia, and Escherichia coli was added during the lysis step of DNA extraction. Phosphate-buffered saline and blank controls were also tested. One 5-μm section of each specimen was extracted and all DNA extracts were tested for interference of mutation detection using the cobas® test.
Cross-reactivity
The cross-reactivity of the cobas® test with a panel of KRAS silent mutations and KRAS homolog plasmids was assessed. Plasmids containing one each of three silent codon 12 mutations and three silent codon 13 mutations were prepared in a background of wild-type cell line DNA and tested in the presence and absence of 5% KRAS mutant DNA. Additionally, cross-reactivity and interference of the cobas® test were assessed with highly homologous RAS gene sequences present in the human genome (HRAS, NRAS, and pseudogene KRAS1P). Six plasmids (KRAS codon 12/13 pseudogene, codon 61 pseudogene, NRAS exon 2, NRAS exon 3, HRAS exon 2, and HRAS exon 3) were blended with KRAS wild-type DNA (K562 cell line DNA) and tested in the presence and absence of 5% KRAS mutant DNA. Each blend was tested in triplicate.
Necrosis
The impact of tissue necrosis on the performance of the cobas® test was evaluated in duplicate in 20 FFPET CRC specimens with varying percentages of necrotic tissue (5–70%) as assessed by an external pathologist. Mutation status was verified by Sanger sequencing. The FFPET specimens used were:
-
Ten wild-type specimens with 30–70% necrotic tissue,
-
Five KRAS codon 12 mutants with 30–45% necrotic tissue,
-
Four KRAS codon 13 mutants with 15–50% necrotic tissue, and
-
One KRAS codon 61 mutant with 5% necrotic tissue.
Results
Limit of detection
For the LOD analysis for the three predominant mutations in codons 12, 13, and 61, ≥95% correct mutation call rates were obtained across all specimen types with approximately 5% mutant alleles (as assessed by 454) at a DNA input of 0.8–6.3 ng per reaction well (Table 1). A 100% correct mutation call rate for the remaining 16 mutations in codons 12, 13, and 61 for all specimens was obtained at the recommended DNA input of 50 ng per reaction well (Table 2).
Methods correlation
The analytic performance of the cobas® test and 2X bi-directional Sanger sequencing was compared with a total of 188 FFPET vendor-purchased specimens using two different cobas® test reagent lots. Concordance between the cobas® test and Sanger sequencing for each lot was 97.5% positive percent agreement (PPA) and 94.4% negative percent agreement (NPA) for codon 12/13 and 85.7% PPA and 99.4% NPA for codon 61 (Table 3, columns A and B). Eight discordant results for codon 12/13 and two for codon 61 were subjected to 454 sequencing. Six specimens were reported mutation-detected codon 12/13 by the cobas® test and mutation not detected by Sanger. 454 results agreed with the cobas® test in five of the six cases. The other two specimens were reported mutation not detected codon 12/13 by the cobas® test and mutation detected by Sanger. 454 agreed with the cobas® test in both cases. Therefore, 454 sequencing results agreed with the cobas® test for seven of the eight discrepant specimens. One unique specimen per lot remained discordant for codon 12/13 mutation status after 454 sequencing. Both discordant specimens displayed low peak heights in the cobas assay, suggesting the presence of very low levels of mutant alleles. After discordant resolution with 454 sequencing, the PPA for codon 12/13 was 100% and the NPA was 99% (Table 3, column C). 454 sequencing results of the two discordant specimens for codon 61 agreed with the cobas® test. After discordant resolution with 454 sequencing, the PPA and the NPA for codon 61 were 100% (Table 3, column D). In addition to the 10 discrepant specimens, 50 specimens with concordant cobas® test and Sanger sequencing results (26 codon 12/13 mutations, 6 codon 61 mutations, and 18 mutation not detected specimens) were subjected to 454 sequencing. The 454 sequencing results for all 50 concordant specimens were concordant with the Sanger sequencing and cobas® KRAS test results.
Repeatability
All 16 repeatability runs were valid. The cobas® test had a mutation result accuracy of 100% (192/192 replicates) across all specimens, reagent lots, operators, and instruments combined (Table 4).
Interfering substances
No interference was observed for any of the 10 specimens tested for potential endogenous interfering substances (hemoglobin and triglycerides) at Clinical and Laboratory Standards Institute recommended test concentrations of 2 g/L and 37 mM, respectively. None of the exogenous colon-related microorganisms tested interfered with the cobas® test. All five of the KRAS wild-type specimens spiked with an exogenous substance gave valid results of “mutation not detected”, while all five KRAS-mutant specimens spiked with an exogenous substance gave valid results of “mutation detected” for the correct codon. Both study controls gave valid and accurate mutation results (data not shown).
Cross-reactivity
The cobas® test did not cross-react with either KRAS silent mutations in codons 12/13 or highly homologous RAS gene sequences. When tested in a wild-type background, all plasmid blends tested with either KRAS silent mutations or KRAS homologs resulted in “mutation not detected”. When tested in a background of 5% KRAS-mutant DNA, all plasmid blends resulted in “mutation detected” for the correct codon (data not shown).
Necrosis
No interference from necrotic tissue was observed when evaluating the performance of the cobas® test. Results for all specimens were concordant with Sanger and 454 sequencing results. Necrosis up to 70% did not affect cobas® test performance (data not shown).
Discussion
KRAS testing is now mandatory for patients with CRC being considered for treatment with anti-EGFR monoclonal antibodies such as cetuximab (Erbitux®, Bristol Myers Squibb and Merck-Serono) and panitumumab (Vectibix®, Amgen). Several methods for KRAS testing currently exist, from commercially available CE-marked test kits to laboratory-developed tests, but there are limited data to support the performance of individual methods over others [21–23]. With the lack of standardization for KRAS testing, there is a widely recognized need for external quality assessment programs and standards [24, 25]. Indeed, a number of recent studies have highlighted the variability of mutation test results from different clinical laboratories [26, 27]. As results directly affect patient therapy, KRAS tests should be thoroughly validated on clinically relevant specimen types (including FFPET) with optimal analytical performance that encompasses a comprehensive understanding of the impact of specimen attributes, such as the levels of amplifiable DNA; the tumor content of FFPET specimens used for analysis; tissue necrosis; and the influence of endogenous and exogenous interfering substances.
The cobas® test was thoroughly evaluated with 23 unique performance verification studies and in 188 CRC clinical specimens. Here, we have reported results on six of these studies, including methods correlation with Sanger sequencing, limit of detection, repeatability, interfering substances, cross-reactivity, and necrosis. These verification tests were performed in FFPET specimens as well as DNA derived from cell lines and plasmids. Specimen attributes that were evaluated were: tumor cell content, potential effects of endogenous and exogenous interfering substances, cross-reactivity with KRAS silent mutations and KRAS homologs, and proportion of necrotic tissue in the FFPET specimen. These analytic performance attributes have not been published for other KRAS testing methods.
Concordance between the cobas® test and sequencing (Sanger plus 454) was 99.5% for codon 12/13 and 100% for codon 61. The cobas® test had a ≥95% correct mutation call rate across all specimen types with ~5% mutant alleles at a DNA input of 0.8–6.3ng/PCR, and a 100% correct mutation call rate across all specimen types for the recommended DNA input of 50 ng per reaction well. The ability of this assay to detect KRAS mutations at lower DNA inputs reduces the likelihood that mutations will be missed in poor-quality FFPET samples with limited amounts of amplifiable DNA. Because we did not perform a systematic assessment of samples with <5% mutant alleles, the detectability of lower levels of mutation with this assay is uncertain.
The cobas® test had a mutation result accuracy of 100% for all specimens and operators combined in the repeatability study. Neither endogenous interfering factors (hemoglobin or triglycerides) nor the presence of exogenous colon-related microorganisms interfered with the test results. The cobas® test does not cross-react with KRAS silent mutations or KRAS homologs from the RAS gene family. The cobas® test gave 100% correct mutation call in FFPET specimens with up to 70% necrosis and ≥95% correct mutation call rate in specimens with low tumor content at ≤50 ng per reaction well.
In contrast to other KRAS testing kits, such as the therascreen® KRAS test (Qiagen, Manchester, UK), which identifies seven somatic mutations in codons 12 (six mutations) and 13 (one mutation), the cobas® test identifies 19 mutations in codons 12, 13, and 61. Based on data from the Catalogue of Somatic Mutations in Cancer (COSMIC) database [10], the additional 12 mutations detected by the cobas test represent approximately 2% of all KRAS mutations or approximately 1% of all CRC cases. However, recent studies suggest that codon 61 mutations are more prevalent than reflected in the COSMIC data [28, 29]. Thus, more comprehensive KRAS mutation coverage may help to improve the selection of patients for this treatment [15]. The analytical studies presented in this paper demonstrate that the cobas® KRAS Mutation Test is a thoroughly validated, robust, and reproducible companion diagnostic test for the detection of the KRAS mutation in CRC tumors from patients under consideration for treatment with anti-EGFR monoclonal antibodies.
References
Bos JL (1989) ras oncogenes in human cancer: a review. Cancer Res 49(17):4682–4689
Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N (2001) Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 85(5):692–696. doi:10.1054/bjoc.2001.1964
Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML (2000) Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev 9(11):1193–1197
Ciardiello F, Tortora G (2008) EGFR antagonists in cancer treatment. N Engl J Med 358(11):1160–1174. doi:10.1056/NEJMra0707704
Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F (2011) Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 29(15):2011–2019. doi:10.1200/JCO.2010.33.5091
Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P (2009) Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 27(5):663–671
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26(10):1626–1634
Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359(17):1757–1765. doi:10.1056/NEJMoa0804385
Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA, Richel DJ, Voest EE, Dijkstra JR, Vink-Borger ME, Antonini NF, Mol L, van Krieken JH, Dalesio O, Punt CJ (2009) Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med 360(6):563–572
Catalog Of Somatic Mutations In Cancer (COSMIC) database, v. 54 (2011) (http://www.sanger.ac.uk/perl/genetics/CGP/cosmic)
Der CJ, Finkel T, Cooper GM (1986) Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 44(1):167–176
Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277(5324):333–338
Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F (2009) KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 101(4):715–721
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11(8):753–762
Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R, Alabiso O, Girlando S, Soini B, Spitale A, Di Nicolantonio F, Saletti P, Crippa S, Mazzucchelli L, Marchetti A, Bardelli A, Frattini M (2011) Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res 17(14):4901–4914. doi:10.1158/1078-0432.CCR-10-3137
Agency EM (2008) Committee for Medicinal Products for Human Use post-authorisation summary of positive opinion for Erbitux., vol doc. Ref
Administration USFaD (2009) cetuximab (Erbitux) and panitumumab (Vectibix)
Van Cutsem E, Oliveira J (2009) Advanced colorectal cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol 20(Suppl 4):61–63. doi:10.1093/annonc/mdp130
Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27(12):2091–2096
Querings S, Altmuller J, Ansen S, Zander T, Seidel D, Gabler F, Peifer M, Markert E, Stemshorn K, Timmermann B, Saal B, Klose S, Ernestus K, Scheffler M, Engel-Riedel W, Stoelben E, Brambilla E, Wolf J, Nurnberg P, Thomas RK (2011) Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One 6(5):e19601. doi:10.1371/journal.pone.0019601
Gao J, Li YY, Sun PN, Shen L (2010) Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol 16(38):4858–4864
Angulo B, Garcia-Garcia E, Martinez R, Suarez-Gauthier A, Conde E, Hidalgo M, Lopez-Rios F (2010) A commercial real-time PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations: a morphology-based approach in colorectal carcinoma. J Mol Diagn 12(3):292–299
Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, Evans TJ, Ismail T, Li WQ, Collins P, Ravetto P, Leggett B, Salto-Tellez M, Soong R, Fox S, Scott RJ, Dobrovic A, Iacopetta B (2009) A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn 11(6):543–552
van Krieken H, Tol J (2009) Setting future standards for KRAS testing in colorectal cancer. Pharmacogenomics 10(1):1–3
Dequeker E, Ligtenberg MJ, Vander Borght S, van Krieken JH (2011) Mutation analysis of KRAS prior to targeted therapy in colorectal cancer: development and evaluation of quality by a European external quality assessment scheme. Virchows Arch 459(2):155–160. doi:10.1007/s00428-011-1094-x
Bellon E, Ligtenberg MJ, Tejpar S, Cox K, de Hertogh G, de Stricker K, Edsjo A, Gorgoulis V, Hofler G, Jung A, Kotsinas A, Laurent-Puig P, Lopez-Rios F, Hansen TP, Rouleau E, Vandenberghe P, van Krieken JJ, Dequeker E (2011) External quality assessment for KRAS testing is needed: setup of a European program and report of the first joined regional quality assessment rounds. Oncologist 16(4):467–478
Beau-Faller M, Degeorges A, Rolland E, Mounawar M, Antoine M, Poulot V, Mauguen A, Barbu V, Coulet F, Pretet JL, Bieche I, Blons H, Boyer JC, Buisine MP, de Fraipont F, Lizard S, Olschwang S, Saulnier P, Prunier-Mirebeau D, Richard N, Danel C, Brambilla E, Chouaid C, Zalcman G, Hainaut P, Michiels S, Cadranel J (2011) Cross-validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non-small cell lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR-TKIs in non-small cell lung carcinoma [ERMETIC] project—part 1). J Thorac Oncol 6(6):1006–1015. doi:10.1097/JTO.0b013e318211dcee
Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P (2009) KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 27(35):5931–5937
Gaedcke J, Grade M, Jung K, Schirmer M, Jo P, Obermeyer C, Wolff HA, Herrmann MK, Beissbarth T, Becker H, Ried T, Ghadimi M (2010) KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol 94(1):76–81
Acknowledgments
We acknowledge Felice Shieh for her contributions to manuscript preparation.
Conflict of interest
All authors are full-time employees of Roche Molecular Systems, Inc., Pleasanton, CA.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lee, S., Brophy, V.H., Cao, J. et al. Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch 460, 141–149 (2012). https://doi.org/10.1007/s00428-011-1180-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-011-1180-0