Abstract
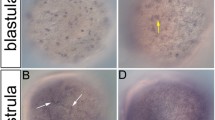
Notch genes encode transmembrane receptors that interact with numerous signal transduction pathways and are essential for animal development. To facilitate analysis of vertebrate Notch gene function, we isolated cDNA fragments of three novel Notch genes from zebrafish (Danio rerio), Notch1b, Notch5 and Notch6. Notch1b is a second zebrafish Notch1 gene. From analysis of the Notch1b sequence we argue that the various vertebrate Notch gene subfamilies encode receptors with different signalling specificities. Notch5 and Notch6 represent novel vertebrate Notch gene subfamilies. Remarkably, Notch1b lacks expression in presomitic mesoderm, Notch5 is expressed in a metameric pattern within the presomitic mesoderm whilst Notch6 expression is excluded from the nervous system. The expression patterns of these genes suggest important roles in gastrulation, somitogenesis, tail bud extension, myogenesis, heart development and neurogenesis. We discuss the implications of our observations for Notch gene evolution and function.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 20 January 1997 / Accepted: 12 February 1997
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Figure S1 A, B: Alignments of putative translations of the novel zebrafish Notch genes against the Notch1a receptor ("zebrafish Notch")
The region of each Notch gene reading frame cloned is indicated relative to a typical Notch family receptor such as Notch1a. The first amino acid residues (aa) of EGF repeats (EGFR), LN repeats (LNR) and ankyrin repeats (ankyrin R) are indicated beneath the Notch1a sequence by asterisks followed by the repeats' names and numbers. Numbers above and below the alignments indicate aa number. The cDNA binding positions for degenerate PCR oligonucleotides used to amplify the different regions of the Notch reading frames are indicated by horizontal arrows. A) Alignment of Notch1a with Notch1b (left) and Notch5 (right). B) Alignment of Notch1a with Notch6. Vertical arrows indicate the boundaries of the EGF repeat region that was deleted in the original Notch6 isolates (see Experimental Procedures). The cDNA sequences have been deposited in the EMBL nucleotide sequence database under accession numbers Y10352 (Notch1b), Y10353 (Notch5) and Y10354 (Notch6).
Figure S1 A
Alignment of Notch1a with Notch1b (left) and Notch5 (right). (PDF 15 KB)
Figure S1 B
Alignment of Notch1a with Notch6 (PDF 14 KB)
Rights and permissions
About this article
Cite this article
Westin, J., Lardelli, M. Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev Gene Evol 207, 51–63 (1997). https://doi.org/10.1007/s004270050091
Issue Date:
DOI: https://doi.org/10.1007/s004270050091