Abstract
Spiralian embryogenesis is deeply conserved and seems to have been in place in the last common ancestor of the large assemblage of protostome phyla known as the Lophotrochozoa. While the blastula fate maps of several spiralian embryos have been determined, little is known about the events that link the early embryo and the larva. For all cells in the Ilyanassa blastula, we determined the clonal morphology at four time points between the blastula and veliger stages. We found that ectomesoderm comes mostly from 3a and 3b, but also from 2c and 2b. We also observed the ingression and early proliferation of 3a- and 3b-derived ectomesoderm. We found cells in the 2b clone that marked the anterior edge of the blastopore and later the mouth and cells in the 3c/3d clones that marked the posterior edges of these structures. This demonstrates directly that the mouth forms in the same location as the blastopore. In the development of the shell field, we observed dramatic cell migration events that invert the positions of the 2b and 2d clones that contribute to the shell. Using time-lapse imaging, we followed and described the cleavage pattern of the conserved endomesodermal blast cell, 4d, up to 4d + 45 h, when there were 52 cells in the clone. Our results show the growth and movement of clones derived from cells of the spiralian blastula as they transform into the trochophore-like and veliger stages. They have implications for the evolution of the shell in gastropods, the origins of mesoderm in spiralians, and the evolution of mouth formation in metazoans.







Similar content being viewed by others
References
Ackermann C, Dorresteijn A, Fischer A (2005) Clonal domains in postlarval Platynereis dumerilii (Annelida:Polychaeta). J Morphol 266:258–280
Arendt D, Nubler-Jung K (1997) Dorsal or ventral: similarities in fate maps and gastrulation patterns in annelids, arthropods and chordates. Mech Dev 61:7–21
Arendt D, Technau U, Wittbrodt J (2001) Evolution of the bilaterian larval foregut. Nature 409:81–85
Boyer BC, Henry JQ (1998) Evolutionary modifications of the spiralian developmental program. Am Zool 38:621–633
Boyer BC, Henry JQ, Martindale MQ (1996) Dual origins of mesoderm in a basal spiralian: cell lineage analyses in the polyclad turbellarian Hoploplana inquilina. Dev Biol 179:329–338
Cather JN (1967) Cellular interactions in the development of the shell gland of the gastropod Ilyanassa. J Exp Zool 166:205–224
Chan XY, Lambert JD (2011) Patterning a spiralian embryo: a segregated RNA for a Tis11 ortholog is required in the 3a and 3b cells of the Ilyanassa embryo. Dev Biol 349:102–112
Clement AC (1960) Development of the Ilyanassa embryo after removal of the mesentoblast cell. Biol Bull 119:310
Clement AC (1962) Development of Ilyanassa following the removal of the D macromere at successive cleavage stages. J Exp Zool 149:193–216
Clement AC (1967) The embryonic value of micromeres in Ilyanassa obsoleta, as determined by deletion experiment. I. The first quartet cells. J Exp Zool 166:77–88
Clement AC (1986a) The embryonic value of the micromeres in Ilyanassa obsoleta, as determined by deletion experiments: 3) The 3rd quartet cells and the mesentoblast cell, 4d. Int J Invertebr Reprod Dev 9:155–168
Clement AC (1986b) The embryonic value of the micromeres in Ilyanassa obsoleta, as determined by deletion experiments. 2. The 2nd quartet cells. Int J Invertebr Reprod Dev 9:139–153
Conklin EG (1897) The embryology of Crepidula, a contribution to the cell lineage and early development of some marine gasteropods. J Morphol 13:1–226
Dictus WJ, Damen P (1997) Cell-lineage and clonal-contribution map of the trochophore larva of Patella vulgata (Mollusca). Mech Dev 62:213–226
Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, Sorensen MV, Haddock SH, Schmidt-Rhaesa A, Okusu A, Kristensen RM, Wheeler WC, Martindale MQ, Giribet G (2008) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749
Ebstein BS, Rosenthal MD, DeHaan RL (1965) Cells from isolated blastomeres of Ilyanassa obsoleta in tissue culture. Exp Cell Res 40:174–177
Gharbiah M, Cooley J, Leise EM, Nakamoto A, Rabinowitz JS, Lambert JD, Nagy LM (2008) The snail Ilyanassa: a reemerging model for studies in development, emerging model organisms. Cold Spring Harbor Press, Cold Spring Harbor
Gharbiah M, Nakamoto A, Nagy LM (2013) Analysis of ciliary band formation in the mollusc Ilyanassa obsoleta. Dev Genes Evol 223:225–235
Goulding MQ (2009) Cell lineage of the Ilyanassa embryo: evolutionary acceleration of regional differentiation during early development. PLoS One 4:e5506
Hejnol A, Martindale MQ, Henry JQ (2007) High-resolution fate map of the snail Crepidula fornicata: the origins of ciliary bands, nervous system, and muscular elements. Dev Biol 305:63–76
Henry JJ, Martindale MQ (1998) Conservation of the spiralian developmental program: cell lineage of the nemertean, Cerebratulus lacteus. Dev Biol 201:253–269
Henry JQ, Okusu A, Martindale MQ (2004) The cell lineage of the polyplacophoran, Chaetopleura apiculata: variation in the spiralian program and implications for molluscan evolution. Dev Biol 272:145–160
Henry JJ, Collin R, Perry KJ (2010) The slipper snail, Crepidula: an emerging lophotrochozoan model system. Biol Bull 218:211–229
Hodor PG, Ettensohn CA (1998) The dynamics and regulation of mesenchymal cell fusion in the sea urchin embryo. Dev Biol 199:111–124
Kanda T, Sullivan KF, Wahl GM (1998) Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 8:377–385
Kingsley EP, Chan XY, Duan Y, Lambert JD (2007) Widespread RNA segregation in a spiralian embryo. Evol Dev 9:527–539
Lambert JD (2008) Mesoderm in spiralians: the organizer and the 4d cell. J Exp Zool B Mol Dev Evol 310:15–23
Lambert JD (2009) Patterning the spiralian embryo: insights from Ilyanassa. In: Jeffery WR (ed) Curr. Top. Dev. Biol. Academic, Burlington, pp 107–133
Lillie FR (1895) The embryology of the Unionidae. A study in cell-lineage. J Morphol 10:1–100
Lyons DC, Perry KJ, Lesoway MP, Henry JQ (2012) Cleavage pattern and fate map of the mesentoblast, 4d, in the gastropod Crepidula: a hallmark of spiralian development. EvoDevo 3:21
Maduro MF (2010) Cell fate specification in the C. elegans embryo. Dev Dyn 239:1315–1329
Martindale MQ, Hejnol A (2009) A developmental perspective: changes in the position of the blastopore during bilaterian evolution. Dev Cell 17:162–174
Meyer NP, Seaver EC (2010) Cell lineage and fate map of the primary somatoblast of the polychaete annelid Capitella teleta. Integr Comp Biol 50:756–767
Meyer NP, Boyle MJ, Martindale MQ, Seaver EC (2010) A comprehensive fate map by intracellular injection of identified blastomeres in the marine polychaete Capitella teleta. EvoDevo 1:8
Nishida H (2002) Specification of developmental fates in ascidian embryos: molecular approach to maternal determinants and signaling molecules. Int Rev Cytol 217:227–276
Page LR (1997) Ontogenetic torsion and protoconch form in the archaeogastropod Haliotis kamtschatkana: evolutionary implications. Acta Zool 78:227–245
Rabinowitz JS, Lambert JD (2010) Spiralian quartet developmental potential is regulated by specific localization elements that mediate asymmetric RNA segregation. Development 137:4039–4049
Rabinowitz JS, Chan XY, Kingsley EP, Duan Y, Lambert JD (2008) Nanos is required in somatic blast cell lineages in the posterior of a mollusk embryo. Curr Biol 18:331–336
Render J (1991) Fate maps of the 1st quartet micromeres in the gastropod Ilyanassa obsoleta. Development 113:495–501
Render J (1997) Cell fate maps in the Ilyanassa obsoleta embryo beyond the third division. Dev Biol 189:301–310
Rivest BR (1992) Studies on the structure and function of the larval kidney complex of prosobranch gastropods. Biol Bull 182:305–323
Rouse GW (1999) Trochophore concepts: ciliary bands and the evolution of larvae in spiralian Metazoa. Biol J Linn Soc 66:411–464
Shimizu K, Sarashina I, Kagi H, Endo K (2011) Possible functions of Dpp in gastropod shell formation and shell coiling. Dev Genes Evol 221:59–68
Sweet HC (1998) Specification of first quartet micromeres in Ilyanassa involves inherited factors and position with respect to the inducing D macromere. Development 125:4033–4044
Tomlinson SG (1987) Intermediate stages of the embryonic development of the gastropods Ilyanassa obsoleta: a scanning electron microscope study. Int J Invertebr Reprod Dev 12:253–280
Wierzejski A (1905) Embryologie von Physa fontinalis L. Z Wiss Zool 83:502–706, plates 518–527
Wilson EB (1892) The cell lineage of Nereis. J Morphol 6:361–480
Wilson EB (1904) Experimental studies in germinal localization. II. Experiments on the cleavage-mosaic in Patella and Dentalium. J Exp Zool 1:197–268
Acknowledgments
We thank Morgan Goulding, Pamela Agbu and Adam Johnson for thoughtful comments on the manuscript. This work was supported by a grant by the N.S.F to J.D.L. (IOB-1146782).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: David A Weisblat
Electronic supplementary material
Below is the link to the electronic supplementary material.
Movie S1
Stack of confocal images of 1a clone at veliger larva stage. Anterior view. (MPG 1,374 kb)
Movie S2
Stack of confocal images of 1c clone at veliger larva stage. Anterior view. (MPG 1,622 kb)
Movie S3
Stack of confocal images of 1a clone at two day-old stage. Dorsal view. (MPG 2,012 kb)
Movie S4
Stack of confocal images of 1c clone at two day-old stage. Dorsal view. (MPG 1,674 kb)
Movie S5
Stack of confocal images of 1a clone at three day-old stage. Dorsal view. (MPG 1,326 kb)
Movie S6
Stack of confocal images of 1c clone at three day-old stage. Dorsal view. (MPG 2,016 kb)
Movie S7
Stack of confocal images of 1a clone at four day-old stage. Dorsal view. (MPG 1,732 kb)
Movie S8
Stack of confocal images of 1c clone at four day-old stage. Dorsal view. (MPG 2,158 kb)
Movie S9
Stack of confocal images of 1b clone at veliger larva stage. Anterior view. (MPG 766 kb)
Movie S10
Stack of confocal images of 1b clone at two day-old stage. Dorsal view. (MPG 1,634 kb)
Movie S11
Stack of confocal images of 1b clone at two day-old stage. Ventral view. (MPG 1,580 kb)
Movie S12
Stack of confocal images of 1b clone at three day-old stage. Dorsal view. (MPG 1,530 kb)
Movie S13
Stack of confocal images of 1b clone at four day-old stage. Dorsal view. (MPG 1,244 kb)
Movie S14
Stack of confocal images of 1d clone at veliger larva stage. Anterior view. (MPG 870 kb)
Movie S15
Stack of confocal images of 1d clone at two day-old stage. Dorsal view. (MPG 1,102 kb)
Movie S16
Stack of confocal images of 1d clone at three day-old stage. Dorsal view. (MPG 1,134 kb)
Movie S17
Stack of confocal images of 1d clone at four day-old stage. Ventral view. (MPG 2,316 kb)
Movie S18
Stack of confocal images of 2a clone at veliger larva stage. Ventral view. (MPG 1,594 kb)
Movie S19
Stack of confocal images of 2c clone at veliger larva stage. Ventral view. (MPG 1,316 kb)
Movie S20
Stack of confocal images of 2c clone at veliger larva stage. Left view. (MPEG 1,109 kb)
Movie S21
Stack of confocal images of 2a clone at two day-old stage. Posterior ventral view. (MPG 2,036 kb)
Movie S22
Stack of confocal images of 2c clone at two day-old stage. Posterior ventral view. (MPG 2,156 kb)
Movie S23
Stack of confocal images of 2a clone at three day-old stage. Ventral view. (MPG 1,358 kb)
Movie S24
Stack of confocal images of 2c clone at three day-old stage. Ventral view. (MPG 2,020 kb)
Movie S25
Stack of confocal images of 2a clone at four day-old stage. Ventral view. (MPG 924 kb)
Movie S26
Stack of confocal images of 2c clone at four day-old stage. Ventral view. (MPG 992 kb)
Movie S27
Stack of confocal images of 2c clone at four day-old stage. Dorsal view. (MPG 1,220 kb)
Movie S28
Stack of confocal images of 2b clone at veliger larva stage. Anterior view. (MPG 1,566 kb)
Movie S29
Stack of confocal images of 2b clone at two day-old stage. Ventral view. (MPG 1,344 kb)
Movie S30
Stack of confocal images of 2b clone at three day-old stage. Dorsal view. (MPG 1,794 kb)
Movie S31
Stack of confocal images of 2b clone at three day-old stage. Ventral view. (MPG 1,390 kb)
Fig. S32
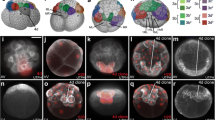
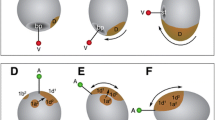
The development of the 2b clone. (A–D) Ventral view. At two days-old, the future mouth cells are positioned on the ventral midline and connected to the band of cells. During the transition from two day-old to three day-old, they disconnected from the band of cells and remain on the ventral midline. (E–H) Dorsal view. At two day-old, the arms of the band of cells are apart from each other on the dorsal side. Transitioning from two day-old to three day-old, the arms of the band of cells extend and join at the dorsal side. In a three day-old embryo, the cells that are part of the shell field bud off from the joined arms. Green: Oregon Green dextran; Blue: DAPI. Dotted line: blastopore; asterisk: developing mouth. (DOCX 322 kb)
Movie S33
Stack of confocal images of 2b clone at four day-old stage. Dorsal view. (MPG 1,154 kb)
Movie S34
Stack of confocal images of 2b clone at four day-old stage. Ventral view. (MPG 1,930 kb)
Movie S35
Stack of confocal images of 2d clone at veliger larva stage. Anterior view. (MPG 908 kb)
Movie S36
Stack of confocal images of 2d clone at two day-old stage. Posterior dorsal view. (MPG 1,224 kb)
Movie S37
Stack of confocal images of 2d clone at three day-old stage. Ventral view. (MPG 2,096 kb)
Movie S38
Stack of confocal images of 2d clone at four day-old stage. Ventral view. (MPG 1,186 kb)
Movie S39
Stack of confocal images of 3a clone at veliger larva stage. Anterior view. (MPG 3,396 kb)
Movie S40
Stack of confocal images of 3b clone at veliger larva stage. Anterior view. (MPG 1,968 kb)
Movie S41
Stack of confocal images of 3a clone at two day-old stage. Ventral view. (MPG 606 kb)
Movie S42
Stack of confocal images of 3b clone at two day-old stage. Ventral view. (MPG 432 kb)
Fig. S43
The mesodermal population derived from the ectomesodermal clone, 3a. (A–B) Ventral view of two day-old embryo. (A) Projection of stacks of mesodermal layers in the two day-old embryo. The mesodermal progenitor cells derived from the 3a clone is shown. (B) Projection of a whole mount embryo showing the clonal distribution of the 3a cell. (C–D) Ventral view of three day-old embryo. (C) Projection of stacks of mesodermal layers in the three day-old embryo. The mesodermal population has divided and extended to the posterior region. Meanwhile, the esophageal cell sheet (ectodermal) has started internalizing through the future mouth opening. (D) Projection of a whole mount three day-old embryo showing the 3a lineage. Green is lineage tracer. Blue is DAPI staining of nuclei. Red is phalloidin staining of filamentous actin. (DOCX 239 kb)
Movie S44
Stack of confocal images of 3a clone at three day-old stage. Ventral view. Red: rhodamine dextran; cyan blue: phalloidin; Blue: DAPI. (MPEG 1,109 kb)
Movie S45
Stack of confocal images of 3b clone at three day-old stage. Ventral view. (MPG 1,128 kb)
Movie S46
Stack of confocal images of 3a clone at four day-old stage. Ventral view. Red: rhodamine dextran; cyan blue: phalloidin; Blue: DAPI. (MPG 2,132 kb)
Movie S47
Stack of confocal images of 3b clone at four day-old stage. Ventral view. (MPG 892 kb)
Movie S48
Stack of confocal images of 3c clone at veliger larva stage. Anterior view. (MPG 1,046 kb)
Movie S49
Stack of confocal images of 3d clone at veliger larva stage. Anterior view. (MPG 746 kb)
Movie S50
Stack of confocal images of 3c clone at two day-old stage. Posterior ventral view. (MPEG 869 kb)
Movie S51
Stack of confocal images of 3d clone at two day-old stage. Posterior ventral view. (MPG 1,350 kb)
Movie S52
Stack of confocal images of 3c clone at three day-old stage. Ventral view. (MPG 3,512 kb)
Movie S53
Stack of confocal images of 3d clone at three day-old stage. Ventral view. (MPG 1,558 kb)
Movie S54
Stack of confocal images of 3c clone at four day-old stage. Ventral view. (MPG 1,120 kb)
Movie S55
Stack of confocal images of 3d clone at four day-old stage. Ventral view. (MPG 2,798 kb)
Movie S56
Stack of confocal images of 3c clone at veliger larva stage. Right view. (MPEG 1,019 kb)
Movie S57
Stack of confocal images of 4A clone at two day-old stage. Ventral view. (MPG 1,058 kb)
Movie S58
Stack of confocal images of 4B clone at two day-old stage. Ventral view. (MPG 1,356 kb)
Movie S59
Stack of confocal images of 4C clone at two day-old stage. Ventral view. (MPG 1,306 kb)
Movie S60
Stack of confocal images of 4A clone at three day-old stage. Ventral view. (MPG 962 kb)
Movie S61
Stack of confocal images of 4B clone at three day-old stage. Ventral view. (MPG 1,870 kb)
Movie S62
Stack of confocal images of 4C clone at three day-old stage. Ventral view. (MPG 1,646 kb)
Movie S63
Stack of confocal images of 4A clone at four day-old stage. Right view. (MPG 1,462 kb)
Movie S64
Stack of confocal images of 4B clone at four day-old stage. Right view. (MPG 1,094 kb)
Movie S65
Stack of confocal images of 4C clone at four day-old stage. Right view. (MPG 1,500 kb)
Movie S66
Stack of confocal images of 4A clone at veliger larva stage. Left view. (MPG 6,466 kb)
Movie S67
Stack of confocal images of 4B clone at veliger larva stage. Left view. (MPG 1,268 kb)
Movie S68
Stack of confocal images of 4C clone at veliger larva stage. Left view. (MPG 1,576 kb)
Movie S69
Stack of confocal images of 4a clone at two day-old stage. Posterior ventral view. (MPG 1,246 kb)
Movie S70
Stack of confocal images of 4b clone at two day-old stage. Posterior ventral view. (MPG 1,204 kb)
Movie S71
Stack of confocal images of 4c clone at two day-old stage. Posterior ventral view. (MPG 1,124 kb)
Movie S72
Stack of confocal images of 4a clone at three day-old stage. Ventral view. (MPG 1,434 kb)
Movie S73
Stack of confocal images of 4b clone at three day-old stage. Ventral view. (MPG 2,014 kb)
Movie S74
Stack of confocal images of 4c clone at three day-old stage. Ventral view. (MPG 1,654 kb)
Movie S75
Stack of confocal images of 4a clone at four day-old stage. Ventral view. (MPG 884 kb)
Movie S76
Stack of confocal images of 4b clone at four day-old stage. Dorsal view. (MPG 1,668 kb)
Movie S77
Stack of confocal images of 4c clone at four day-old stage. Ventral view. (MPG 1,058 kb)
Movie S78
Stack of confocal images of 4a clone at veliger larva stage. Right view. (MPG 1,044 kb)
Movie S79
Stack of confocal images of 4b clone at veliger larva stage. Right view. (MPG 942 kb)
Movie S80
Stack of confocal images of 4c clone at veliger larva stage. Left view. (MPG 2,228 kb)
Movie S81
Stack of confocal images of 4d clone at two day-old stage. Posterior dorsal view. (MPG 1,968 kb)
Movie S82
Stack of confocal images of 4d clone at three day-old stage. Ventral view. (MPG 2,226 kb)
Movie S83
Stack of confocal images of 4d clone at four day-old stage. Ventral view. (MPG 4,234 kb)
Movie S84
Stack of confocal images of 4d clone at veliger larva stage. Dorsal view. (MPG 2,800 kb)
Fig. S85
4d blast cell lineage. The reference lineage is the most extended lineage obtained and thus, is used as reference for comparison to other lineages examined. Lineages after the 4d + 24 h mark are derived from the live-time imaging result of four different embryos including the reference embryo. Left axis is time after the birth of 4d. Color code: long orange line (t = 4d + 24 h mark); black lines (reference); red, blue and green lines each represents division happened in three other individual lineages examined. (DOCX 98 kb)
Time-lapse movie of cell divisions in the 4d lineages. This time-lapsed movie was recorded at 4d + 26.5 h when each side of the lineage contains 12 cells. Chromatin is visualized with H2B-eGFP. Dorsal view. (MPG 5,794 kb)
Fig. S87
The development of the foregut: mouth and esophagus. The positions of the 2nd quartet cells (2a and 2b) and 3rd quartet cells (3a) in relation to the blastopore and the developing mouth. (A–B) Double labeling of the 2a (green) and 3a (red) clones in two day-old and three day-old embryos. (C–D) Double labeling of the 2b (green) and 3a (red) clones in two day-old and three day-old embryos. Green: Oregon Green dextran; Red: Rhodamine dextran; Blue: DAPI. Dotted line: blastopore/developing mouth. (DOCX 219 kb)
Movie S88
Stack of confocal images of 2a and 3a double labeling at two day-old stage. Ventral view. Red: Rhodamine dextran (3a); Green: Oregon green dextran (2a); cyan blue: phalloidin; Blue: DAPI. (MPEG 3,374 kb)
Movie S89
Stack of confocal images of 2a and 3a double labeling at three day-old stage. Ventral view. Red: Rhodamine dextran (3a); Green: Oregon green dextran (2a); cyan blue: phalloidin; Blue: DAPI. (MPEG 1,198 kb)
Movie S90
Stack of confocal images of 2b and 3a double labeling at two day-old stage. Ventral view. Red: Rhodamine dextran (3a); Green: Oregon green dextran (2b); cyan blue: phalloidin; Blue: DAPI. (MPEG 923 kb)
Movie S91
Stack of confocal images of 2b and 3a double labeling at three day-old stage. Ventral view. Red: Rhodamine dextran (3a); Green: Oregon green dextran (2b); cyan blue: phalloidin; Blue: DAPI. (MPEG 1,241 kb)
Fig. S92
Shell field invagination in early trochophores. (A–D) Individual labeling of all 2nd quartet micromeres. (A) 2a clone contributes to upper left part of the shell field. (B) 2c clone contributes to upper right part of the shell field. (C) Higher magnification of the 2b clone contribution to the upper middle part of the shell gland. (D) 2d clone gives rise to a portion of the ventral part of the shell gland that is similar in size to the 2a and 2c contributions. Green: Oregon Green dextran; Blue: DAPI. (DOCX 220 kb)
Fig. S93
Summary of the early cell lineages and the contributions derived from fate mapping. Abbreviation: L, Left; R, Right. (DOCX 340 kb)
Rights and permissions
About this article
Cite this article
Chan, X.Y., Lambert, J.D. Development of blastomere clones in the Ilyanassa embryo: transformation of the spiralian blastula into the larval body plan. Dev Genes Evol 224, 159–174 (2014). https://doi.org/10.1007/s00427-014-0474-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-014-0474-z