Abstract
The remarkable conservation of Hox clusters is an accepted but little understood principle of biology. Some organizational constraints have been identified for vertebrate Hox clusters, but most of these are thought to be recent innovations that may not apply to other organisms. Ironically, many model organisms have disrupted Hox clusters and may not be well-suited for studies of structural constraints. In contrast, the red flour beetle, Tribolium castaneum, which has a long history in Hox gene research, is thought to have a more ancestral-type Hox cluster organization. Here, we demonstrate that the Tribolium homeotic complex (HOMC) is indeed intact, with the individual Hox genes in the expected colinear arrangement and transcribed from the same strand. There is no evidence that the cluster has been invaded by non-Hox protein-coding genes, although expressed sequence tag and genome tiling data suggest that noncoding transcripts are prevalent. Finally, our analysis of several mutations affecting the Tribolium HOMC suggests that intermingling of enhancer elements with neighboring transcription units may constrain the structure of at least one region of the Tribolium cluster. This work lays a foundation for future studies of the Tribolium HOMC that may provide insights into the reasons for Hox cluster conservation.
Similar content being viewed by others
Introduction
Hox clusters arose near the origins of the animal kingdom (Larroux et al. 2007; Ryan et al. 2007). The last common ancestor of the protostomes and deuterostomes is thought to have had a cluster of at least seven genes characterized by a common transcriptional orientation and by colinearity in the order of the genes and their expression domains along the anterior–posterior axis (reviewed in Garcia-Fernandez 2005).
In various metazoan lineages, Hox clusters have gained or lost genes by duplication and deletion but often have maintained their chromosomal order, transcriptional orientation, and both spatial and temporal colinearity of expression patterns (reviewed in Ferrier and Minguillon 2003), suggesting that Hox cluster organization has been subject to strong constraints during evolution. Classical model systems, such as Drosophila and Caenorhabditis elegans, have provided many important insights into the developmental functions of Hox genes but do not provide particularly good examples of Hox cluster conservation. The Hox cluster of Drosophila melanogaster is split into two parts (the Antennapedia (ANTC) and bithorax (BXC) complexes), shows changes in transcriptional orientation of some genes, and includes interspersed genes of independent origin as well as Hox-derived genes that have evolved novel developmental roles (reviewed in Ferrier and Minguillon 2003). These alterations suggest that the constraints keeping the Hox cluster intact may have been lost in the lineage leading to Drosophila. Additional Hox cluster rearrangements (breaks, microinversions, and gene transpositions) have been found in other Drosophila species (Negre et al. 2003; Negre and Ruiz 2007; Von Allmen et al. 1996) as well as in the silk moth Bombyx mori (Yasukochi et al. 2004). The Hox genes of C. elegans (reviewed in Aboobaker and Blaxter 2003) and the tunicate Oikopleura dioica (Seo et al. 2004) have undergone even more extreme loss and rearrangement such that none of their remaining Hox genes are clustered. In most cases, the Hox genes of these organisms still show spatial but not temporal colinearity. Rapid development seems to be the common denominator among most of these organisms, perhaps making temporal colinearity of Hox genes unnecessary, or even undesirable (Ferrier and Holland 2002; Ferrier and Minguillon 2003; Negre et al. 2005).
While studies of disrupted Hox clusters have provided some insights into Hox cluster maintenance, a more complete understanding will require analysis of organisms where they are still intact. Studies of vertebrate Hox clusters have uncovered several potential mechanisms that may promote temporal colinearity and therefore constrain the organization of these clusters (reviewed in Kmita and Duboule 2003). These include progressive changes in chromatin state along the length of the cluster, varying affinity of regulatory elements to a gradient of signal, and the presence of global enhancer elements outside the cluster that regulate multiple genes within the cluster. However, it is not clear whether these mechanisms apply to other organisms.
Duboule (2007) has suggested that the modern vertebrate Hox clusters are actually more organized than the ancestral cluster. Some of the mechanisms constraining the organization of vertebrate Hox clusters likely evolved concomitant with the co-option of Hox genes for functions such as limb development (Duboule 2007; Kmita and Duboule 2003) and, therefore, may not be applicable to other lineages. Based on this model, we might expect to gain a better understanding of the ancestral constraints on Hox clusters by studying a less organized but still intact cluster. Such clusters have been described in organisms as diverse as the cephalochordate amphioxus (Garcia-Fernandez and Holland 1994; Minguillon et al. 2005), sea urchins (Cameron et al. 2006), and the insects Apis (Honey Bee Genome Sequencing Consortium 2006; Dearden et al. 2006) and Anopheles (Holt et al. 2002; Negre and Ruiz 2007). Evidence also suggests that the red flour beetle, Tribolium castaneum, has an intact Hox cluster. Conventional cloning and sequencing of the portion of the cluster corresponding to the Drosophila Antennapedia complex has shown that this region of the homeotic complex (HOMC) is intact in Tribolium (Brown et al. 2002). Genetic mapping also suggests that the integrity of the Tribolium Hox cluster has been maintained (Beeman 1987). Moreover, the genetic methodologies possible with Tribolium, as well as the application of RNAi, have provided a comprehensive description of the full repertoire of Hox genes and their functions (e.g., Beeman et al. 1993; Beeman et al. 1989; Brown et al. 2000; Shippy et al. 2000; Shippy et al. 2006; Stuart et al. 1991; Stuart et al. 1993; Tomoyasu et al. 2005). The Tribolium genome has recently been sequenced, giving us the opportunity to explore the structure of its Hox cluster in detail. Here, we present an analysis of several Hox mutations along with the transcriptional profile of the cluster during embryonic development. We discuss these results with respect to potential mechanisms of Hox cluster organization and constraint.
Materials and methods
Sequence and transposable element analysis
Sequence analysis was performed using Vector NTI Advance 10 (Invitrogen). Basic Local Alignment Search Tools (BLASTs) against Tribolium genome sequence (Tcas_2.0) were performed at http://www.hgsc.bcm.tmc.edu/blast/blast.cgi?organism= Tcastaneum or http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid= 7070, and subsequent analysis was performed using Genboree (http://www.genboree.org/java-bin/login.jsp) or NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/). The entire HOMC sequence was used as a BLASTn query against a collection of expressed sequence tags (ESTs) provided by Dr. Yoonseong Park (Department of Entomology, Kansas State University, Manhattan, KS, USA). Transposable elements were identified and classified using Censor to search the arthropod subset of Repbase (Kohany et al. 2006).
Array design and probe synthesis
Sequence for the Tribolium HOMC was taken from the Tcas_2.0 Baylor HSGC assembly. The tiled region consists of ∼810,000 bases from LG2 (2,290,000 to 301,000) stretching between the two non-Hox genes flanking the complex. NimbelGen designed ∼50 mer oligos covering this region at two densities: (1) one feature per 91 bp and (2) one feature per 70 bp. An additional region spanning 5 kb on either side of the putative homolog of dme-miR-iab-4 was tiled at a higher density of one feature every 10 bp. Visualization and scaling of tiling data was performed using Integrated Genome Browser (Affymetrix, http://www.affymetrix.com/support/developer/tools/download_igb.affx).
Tribolium 0–72-h-old embryos, grown at 30°C in standard media, were collected by sieving, dechorionated for 2 min in 100% bleach, and homogenized in 200 μl of Trizol using a teflon pestle. Total RNA was then extracted using the standard Trizol protocol (Invitrogen). dsDNA was prepared from ∼10 μg of total RNA with random hexamers according to Kapranov et al. (2002), with the following modifications. Primers were annealed using a 20-min ramp to 15°C, and the first strand reaction was not subdivided for second strand synthesis. The resulting cDNA was used as template by NimbelGen for labeling and hybridization (Squazzo et al. 2006).
Fluorescent in situ hybridization
Probe labeling, embryo fixation, and RNA FISH were performed according to Kosman et al. (2004) with the following modifications. Tribolium embryos were dechorionated in 100% bleach for 2 min and agitated for 45 min to 1 h. Embryos were devitellinized by alternating 1 min vortexing with 1 min shaking for 5 min after the addition of cold methanol, followed by passage through an 18-gauge syringe three to five times. Primers used for making the TcNC-1 and iab-4 probes are as follows: TcNC5′:AGATAAGATATAATGAGGTGTAGAGTTG, TcNC3′: TGATTAACATGGACGGCTTCATTAG, iab-45′: CATCCTATGCACATGCGTTC, iab-43′: CGTTTTAATGGGTGCATCGT. Dig-labeled RNA probes were detected using sheepαDIG (Roche) primary and donkeyαsheep Alexa Fluor 555 (Molecular Probes) secondary antibodies.
Genetics
Beetles were cultured at 30°C on whole wheat flour supplemented with 5% brewer’s yeast as described by Beeman et al. (1989). Strains used were: Ga-1 and Ga-2 (wild type); mxp Dch-3/Ey; Cx 6/A Es; ptl KT76/+; Cx 61/A Es; ptl D60/Ey and Dfd 1/A Es. Eyeless (Ey; Beeman et al. 1996) and Abdominal Extra sclerite (A Es; Beeman et al. 1989) are dominantly marked balancer chromosomes that suppress crossing over within the HOMC.
Cuticle preparations were performed as described by Shippy et al. (2000). For documentation, cuticles were placed in 9:1 lactic acid/ethanol on a depression slide and covered with a coverslip. Images were captured at several focal planes using a Nikon DXM1200F digital camera and combined into a single image using Auto-Montage software (Syncrosopy).
RNAi
Parental RNAi for ptl/Tc-Antp was performed by injection of dsRNA into the abdomens of female pupae. Eggs were collected from injected females at 3-day intervals, aged to hatching, and subjected to cuticle preparation.
Analysis of ptl D60 breakpoints
Eggs were collected after overnight incubation at 30°C and allowed to develop for 3 days. Genomic DNA was isolated from individual ptl D60 homozygous larvae as described by Gloor et al. (1993).
Polymerase chain reaction (PCR) surveys of the HOMC were used to identify likely breakpoint positions, and fragments spanning these putative breakpoints were amplified using universal PCR (Beeman and Stauth 1997; Sarkar et al. 1993). PCR products were cloned and sequenced at the Kansas State University DNA Sequencing Center. The resulting sequences were compared to the Tribolium genome sequence to characterize the breakpoints. GenBank accession numbers for these sequences are as follows: ptlD60 A (EF591668) and ptlD60 B (EF591669).
Analysis of mxp Dch-3 breakpoints
Tribolium genomic DNA was isolated from mxp Dch-3/Ey, A Es/Ey, and Ga-1 pupae as described by Brown et al. (1990), with the exception that DNA was not purified on a CsCl gradient. Digested DNAs were separated on a 0.7% agarose gel by field inversion gel electrophoresis and transferred to GeneScreen nylon membrane (NEN Life Sciences). To look for restriction fragment length polymorphisms associated with mxp Dch-3, the blot was probed with pBmxp2.1, a 5.2 kb HindIII fragment containing the 5′ end of the mxp/Tc-pb coding region.
Inverse PCR (Ochman et al. 1988) was used to clone breakpoints associated with mxp Dch-3. mxp Dch-3/Ey genomic DNA was digested with a restriction enzyme (EcoRI, HindIII, and RsaI for breakpoint fragments A, B, and C, respectively). After circularization of the fragments, two rounds of PCR were performed with primers designed from known sequence. Resulting fragments were cloned using the TOPO-TA Cloning Kit (Invitrogen), sequenced, and submitted to GenBank under the following accession numbers: Dch3 A (EF591670), Dch3 B (EF591671), Dch3 C (EF591672). Sequences were compared to the Tribolium genome sequence to determine the location of breakpoints.
Analysis of ptl KT76 transposon insertion site
The ptl KT76 piggyBac-insertion site was amplified by vectorette PCR as described by Lorenzen et al. (2007). The resulting product was sequenced by Elim Biopharmaceuticals, Inc. (Hayward, CA, USA) and the sequence was submitted to GenBank as accession number EU056827.
Results
The Tribolium Hox cluster has retained an ancestral organization
Several bacterial artificial chromosome (BAC) clones encompassing the ANTC-like region of the Tribolium Hox cluster were previously sequenced and annotated (Brown et al. 2002). Using the newly assembled Tribolium genome sequence, we have performed a similar analysis of the BXC-like portion of the cluster and find that this region contains the Tribolium orthologs of Ultrabithorax (Ubx; Bennett et al. 1999), abdominal-A (abd-A; Shippy et al. 1998) and Abdominal-B (Abd-B). As in Drosophila, the transcription units in this part of the complex are larger than those of the ANTC-like portion due to the presence of longer introns.
As expected from previous molecular and genetic studies (Beeman 1987; Brown et al. 2002), all of the Tribolium Hox genes map to a single cluster on LG2. This cluster spans approximately 756 Kb within a single scaffold of the assembled genome sequence. A few small sequencing gaps are present in the assembly, but more than half can be filled by other available sequences (i.e., the three BAC clones previously sequenced for the ANTC-like portion of the cluster and four BACs from the BXC-like region sequenced for verification of the shotgun genome assembly; Tribolium Genome Consortium 2008). The total length of the filled gaps is approximately 2,635 bp (mismatches in the sequence flanking the gaps lead to some ambiguity), which is only slightly longer than the estimated total length of these gaps (1,938 bp). Thus, estimation of sequencing gaps in the HOMC region appears to be quite accurate.
Two of these gaps are immediately adjacent to transposable element insertion sites and may result from difficulties in assembling repetitive DNA. These two sites account for about 1,300 bp of the total gaps in the HOMC. In two other cases, gaps in the genome assembly are associated with tandem duplications that are not present in the BAC assemblies: an approximately 160-bp duplication between Tc-Deformed (Tc-Dfd) and Tc-zen1 and an approximately 8.5-kb duplication between prothoraxless/Tc-Antennapedia (ptl/Tc-Antp) and Tc-fushi tarazu (Tc-ftz). We designed primers to amplify across the regions in question using Ga-2 genomic DNA (the same inbred strain that was used for the Tribolium genome sequence). In both cases, the size of the resulting fragment is consistent with that predicted from the BAC sequence (data not shown), suggesting that these gaps and duplications are artifacts of the genome assembly process. It is important to note that these artifacts affect only a small fraction of the HOMC sequence, but they underscore the increased quality of finished versus draft sequences.
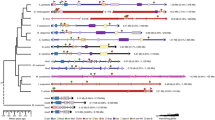
The single Tribolium Hox cluster contains orthologs of all eight Drosophila Hox genes, as well as orthologs of the Hox-derived genes, fushi tarazu and zerknüllt (zen; Tribolium Genome Consortium 2008 and Fig. 1). (In the case of zen, Tribolium has two paralogs apparently resulting from a recent duplication in the beetle lineage, independent of the zen duplication that occurred in the Drosophila lineage (Brown et al. 2002)). These genes are arranged in the same order on the chromosome as their counterparts in other insects. As in Apis (Dearden et al. 2006; Negre and Ruiz 2007), but in contrast to Drosophila and Anopheles (Negre and Ruiz 2007), the Hox and Hox-derived genes in the Tribolium cluster are all oriented in the same direction (Fig. 1). In addition, the two miRNA genes (miR-10 and miR-iab-4) that have been described in other insect Hox clusters are found at conserved positions in the Tribolium HOMC (Tanzer et al. 2005 and Fig. 1).
Embryonic transcription across the complete Tribolium Hox complex. The tiling array consists of ∼50,000 50 bp probes that estimate degree of transcription. Relative intensities for each probe are represented as peaks correlated with a consensus annotation of the Tribolium Hox complex (below). Peak height, shown as Percentile Probe Intensity (PPI), corresponds to the level of transcription for a particular probe. The nucleotide position for each segment is displayed in the upper left and right corners of the panel (numbers correspond with linkage group 2, release Tcas_2.0). New ESTs (cyan) are displayed in the annotation track along with transposable elements (gray). For annotated genes and ESTs, the arrow indicates the direction of transcription. Red arrows indicate the location of two RNA-FISH probes
The ANTC and BXC clusters of Drosophila melanogaster contain a number of non-Hox, protein-coding genes. In contrast, there is no evidence for non-Hox, protein-coding genes in the Tribolium HOMC (The Tribolium Genome Consortium 2008). Here, we corroborate those findings by using several methods to address whether unrelated genes might be interspersed among the Tribolium Hox genes. First, we searched the Tribolium genome for orthologs of genes that are located within the D. melanogaster clusters and determined that none of these genes are located within the Tribolium HOMC. Second, we analyzed predicted proteins within the region to determine whether any of them have recognizable orthologs in other species. Other than the Hox and Hox-derived genes, we found no evolutionarily conserved proteins among either the GLEAN predictions or the GNOMON ab initio predictions that map to the Hox cluster. Third, we searched a collection of Tribolium ESTs for expressed sequences within the HOMC. By this approach, we identified three non-Hox EST clusters that appear to represent noncoding transcripts as well as evidence for a mariner transposase gene (see below), but no other protein-coding genes were found. Finally, we analyzed the embryonically transcribed sequences identified by a tiling array to determine if any were likely to encode proteins. Again, we found no evidence of non-Hox-related protein-coding genes other than those within transposable elements. Although there are caveats to these analyses (e.g., gene prediction methods are imperfect, the tiling array represents only the embryonic transcriptome and EST coverage is incomplete), our results strongly suggest that the protein-coding genes in the Tribolium Hox complex (excluding genes within transposable elements) are all either Hox or Hox-derived genes.
Comparison of transposable element density in the Hox complexes of various animals has led to the suggestion that higher abundance of transposable elements in Hox clusters is correlated with loss of structural integrity. Mammalian Hox complexes have a reduced number of transposons compared to other regions of the genome (Ferrier and Minguillon 2003). Moreover, when transposons are present, they seem to be preferentially inserted into nontranscribed regions of the clusters (Mainguy et al. 2007). In contrast, transposons occur fairly frequently in the split Drosophila clusters (Fried et al. 2004). Though the prediction of three transposable elements in the Tribolium Hox complex (Fig. 1) may be an underestimate, the same method predicts fivefold more in the Drosophila Hox clusters. Additionally, only three transposable elements (all mariners) have been found in the larger but intact Apis Hox complex (Dearden et al. 2006). These numbers are consistent with the apparent inverse correlation between transposon number and the level of Hox cluster organization.
Taken together, these observations suggest that with respect to gene content, order, and orientation, the Tribolium Hox cluster closely resembles the putative ancestral Hox cluster (Garcia-Fernandez 2005). Thus, the constraints preserving the integrity of the Hox cluster may still be in force in Tribolium.
To determine whether these constraints extend outside the Tribolium Hox cluster, we examined synteny beyond the cluster itself. As previously described, Tc-chaoptic partially overlaps the 3′ UTR of Tc-labial (Tc-lab) on the opposite strand (Nie et al. 2001). Working outward, the first gene on the same strand as the Hox genes is Tc_00927, a dolichyl glycosyltransferase orthologous to D. melanogaster CG4542. Beyond the Tc-Abd-B locus is a cluster of putative serine carboxypeptidase genes (Tc_00887, Tc_00664, Tc_00665, and Tc_00666) and the ortholog of D. melanogaster CG3909 (Tc_00886). We identified the orthologs of these genes in D. melanogaster, Anopheles gambiae, and Apis mellifera and determined their map positions. None of these genes map near the Hox cluster in any of the other insects. Likewise, orthologs of the genes adjacent to lab and Abd-B in D. melanogaster do not map near the Hox clusters of the other three insects. These results suggest that the constraints preserving the Hox cluster act only on the Hox genes themselves and not the surrounding region.
The Tribolium Hox cluster produces numerous noncoding transcripts
We developed a tiling microarray covering the Tribolium Hox complex to identify the transcription units active during a broad window of Tribolium embryonic development. Tiling array signal intensity profiles were compared with previously described Hox cDNA structures. Though the tiling density is not fine-scaled enough to effectively resolve intron–exon boundaries, there is a near-perfect correlation between tiling array-predicted transcription and the position of exons in the well-characterized Hox genes. The only caveat is that the 5′ exons of maxillopedia/Tc-proboscipedia (mxp/Tc-pb) and Tc-Abd-B exhibit weaker signals than the other exons of these genes. The most likely explanation is that the 5′ exon is present only in a small subset of the transcripts derived from the gene (i.e., a minor spliceoform).
During the first 3 days of development, numerous regions of the Hox complex, including intergenic and intronic noncoding regions, are actively transcribed (Fig. 1). Interestingly, neither of the two most likely noncoding candidates, the previously described miRNAs, is robustly identified on the tiling array. Transcription at the tca-miR-10 locus is not detected, and transcription at the tca-miR-iab-4 locus is weak. It may be that tca-miR-10 is not expressed during the stages examined, whereas in situ hybridization assays show that tca-miR-iab-4 is strongly expressed during part of the developmental window examined (Fig. 2b).
Expression pattern of two HOMC noncoding transcripts in Tribolium embryos. Probe positions are shown in Fig. 1. a Expression pattern from a 1-kb probe located ∼86 kb 5′ of the start of ptl/Tc-Antp. b The expression pattern of the Tribolium homolog of the iab-4 miRNA (tca-miR-iab-4)
The most intense hybridization signals are detected in the central 250 kb of the Hox complex, encompassing the ptl/Tc-Antp and Ultrathorax/Tc-Ultrabithorax (Utx/Tc-Ubx) genes. Strikingly, transcription in this region is almost equally intense for coding and noncoding loci, and for both the introns and exons of the protein-coding genes. There are hundreds of discrete regions (500 bp or longer) where signal intensity is many times greater than for verified Hox gene exons. It is not possible to determine from the single time point we have analyzed if any of these discrete regions are part of larger transcripts. To verify that the observed signals in the tiling array represent authentic transcription, RNA fluorescent in situ hybridization (FISH) was performed with a representative 1-kb region between ptl/Tc-Antp and Utx/Tc-Ubx (TcNC-1 in Fig. 1). This region is expressed in a Hox-like pattern with distinct anterior and posterior borders in the posterior region of the elongating germ band (Fig. 2a). Interestingly, signal is detected primarily in two spots per nucleus, presumably at the sites of nascent transcription. This suggests either rapid degradation or processing of a primary transcript as would be seen for a pri-miRNA or an intron.
In our search for additional genes within the Tribolium Hox cluster, we identified three ESTs that appear to represent noncoding transcripts. One seems to be a chimeric artifact, arising from the fusion of a tca-miR-10 precursor and part of a 28s rRNA gene. The second, represented by two independent cDNAs, maps between ptl/Tc-Antp and Utx/Tc-Ubx while the third is located within the first intron of Utx/Tc-Ubx. The last two are transcribed from the strand opposite the Hox genes and are correlated with regions of strong signal in the tiling array analysis.
The mxp Dch-3 mutation affects regulation of both mxp/Tc-pb and Cx/Tc-Scr
Because complex regulatory regions may act as a constraining force keeping Hox clusters intact, we analyzed the mxp Dch-3 mutation, which was shown to have unusual effects on the expression of mxp/Tc-pb (Shippy et al. 2000); mxp Dch-3 homozygotes lack most, if not all, normal mxp/Tc-pb expression, but both heterozygotes and homozygotes display strong ectopic mxp/Tc-pb expression in a pattern reminiscent of Cephalothorax/Tc-Sex combs reduced (Cx/Tc-Scr) expression (albeit with an apparent posterior shift of some domains). This ectopic expression is sufficient to rescue some aspects of mxp/Tc-pb function so mxp Dch-3 is not an mxp/Tc-pb null (Shippy et al. 2000). Interestingly, we find that mxp Dch-3 fails to complement a null allele of Cx/Tc-Scr (and, in fact, appears to be null for Cx/Tc-Scr) but complements null alleles of Tc-Dfd and ptl/Tc-Antp (data not shown).
To better understand this complex mutation, we characterized the breakpoints associated with mxp Dch-3. mxp Dch-3 is associated with a chromosomal rearrangement involving at least four breakpoints. Although we have not ruled out the presence of additional breakpoints, the simplest interpretation of our data is that a fragment of the HOMC (including Tc-zen1, Tc-zen2, Tc-Dfd, Cx/Tc-Scr, and Tc-ftz) has been removed from the HOMC and inserted between a fragment of LG9 and a non-HOMC fragment of LG2 (Fig. 3a). This scenario is consistent with previously reported pseudo-linkage of LG2 and LG9 associated with mxp Dch-3 (Beeman et al. 1996) and provides an explanation for the mid-embryonic lethality of the mxp Dch-3 homozygotes (Shippy et al. 2000). That is, non-HOMC breakpoints interrupt both the TFIIA-L ortholog, which is a component of the transcriptional machinery (Yokomori et al. 1993), and a homolog of groucho, which is an important transcriptional corepressor in Drosophila (Jimenez et al. 1997; Paroush et al. 1994). We conclude that the breakpoint between Tc-ftz and ptl/Tc-Antp is likely to account for the loss of Cx/Tc-Scr function associated with mxp Dch-3, probably by separating the Cx/Tc-Scr transcription unit from some or all of its regulatory units. The rearrangement probably juxtaposes these regulatory elements with the mxp/Tc-pb transcription unit, providing a likely explanation for the Cx/Tc-Scr-like expression of mxp/Tc-pb in mxp Dch-3 embryos.
Rearrangements of the HOMC. In these schematic diagrams, the positions of cloned breakpoint fragments are underlined. Wild-type chromosomal position (not to scale) on LG2 (purple) is indicated by a gradient of color to illustrate the effects of inversions. a In the mxp Dch-3 rearrangement, an approximately 150-kb fragment of the HOMC has been transposed between fragments of LG9 and LG2. b The ptl D60 mutation is a large inversion that splits the Hox cluster into two parts. Small fragments at each end of the inversion appear to have been deleted, including part of the ptl/Tc-Antp locus
Cx/Tc-Scr regulatory elements map near or within ptl/Tc-Antp
Beeman et al. (1993) observed that mutations in Cx/Tc-Scr and ptl/Tc-Antp often partially fail to complement one another. Because this is precisely the type of genetic interaction that would be predicted if the separation of Hox genes has deleterious effects, we decided to analyze ptl D60, an allele of ptl/Tc-Antp that shows such effects. The ptl D60 mutation, which results in transformation of the larval legs toward antennae as well as reductions of some labial and thoracic tissue, has been proposed to be a null allele of ptl/Tc-Antp (Beeman et al. 1993). However, adults transheterozygous for ptl D60/ptl D2 have a very different phenotype from that produced by larval ptl/Tc-Antp RNAi (Tomoyasu et al. 2005) and, instead, resemble adults in which both ptl/Tc-Antp and Cx/Tc-Scr have been knocked down (Tomoyasu, personal communication). This result raises the possibility that the ptl D60 mutation affects the function of both genes. To address this issue, we performed parental RNAi with ptl/Tc-Antp and found that the resulting larvae (Fig. 4b) show a phenotype almost identical to that of ptl D60 homozygotes (Fig. 4c), suggesting that the ptl D60 mutation primarily affects the function of ptl/Tc-Antp during embryonic development. However, there is a subtle difference between the ptl D60 and ptl/Tc-Antp RNAi phenotypes in the positioning of the T1 appendages, which are located near the ventral midline in the RNAi larvae but more laterally in the mutants. This difference is likely attributable to partial loss of Cx/Tc-Scr function in ptl D60 because Cx/Scr is responsible for the midline position of the labial appendages in Tribolium (Shippy et al. 2006) and other insects (Hughes and Kaufman 2000; Pattatucci et al. 1991; Rogers et al. 1997). Thus, ptl D60 appears to be not only a null allele of ptl/Tc-Antp but also a hypomorphic allele of Cx/Tc-Scr.
Cuticle and enhancer trap phenotypes of ptl/Tc-Antp mutations. The antennae (ant) and the labial (lab) and thoracic (T1–T3) segments are denoted where relevant. a–e Cuticle preps displaying the phenotypes of wild-type (Ga-1; a), ptl/Tc-Antp RNAi (b), ptl D60/ptl D60 (c), ptl KT76/ptl KT76 (d), and ptl KT76/ptl D60 (e) first instar larvae. Enhancer trap-driven EGFP expression in a ptl KT76 embryo (f) appears in a very similar pattern to Cx/Tc-Scr expression (purple) in a wild-type embryo (g). A ptl KT76 larva (h) and pupa (i) also display EGFP enhancer trap expression in parts of the labial and first thoracic segments
To understand why the ptl D60 mutation affects both ptl/Tc-Antp and Cx/Tc-Scr, we characterized its mutant lesion(s). We found that ptl D60 is associated with an inversion of about 6.6 Mb, with breakpoints in ptl/Tc-Antp and a distant region of LG2 (Fig. 3b). In addition, there are small deletions at each end of the inversion (approximately 3.1 kb of the ptl/Tc-Antp transcription unit including all of exon 2 and approximately 1.5 kb at the other end). Consistent with this conclusion, we find that ptl D60 can act as a crossover suppressor for LG2, reducing recombination between Reindeer (a mutation near one end of LG2) and the HOMC from its normal value of 35–40 cM (Beeman et al. 1996) to approximately 20 cM. These results suggest that breakpoints within the ptl/Tc-Antp gene affect the function of both ptl/Tc-Antp and Cx/Tc-Scr, probably by disrupting the function of Cx/Tc-Scr regulatory elements (see “Discussion”).
Additional evidence for the presence of Cx/Tc-Scr regulatory elements in the vicinity of ptl/Tc-Antp comes from a piggyBac-insertion line recovered during an insertional mutagenesis project. The KT076 line carries a homozygous lethal insertion in the last intron of ptl (Fig. 5). Crosses between KT076 heterozygotes produce a class of embryos (putative homozygotes) with the T1 and T2 legs partially transformed toward antennae (Fig. 4d), a phenotype consistent with partial loss of Ptl/Tc-Antp function. However, individuals carrying the insertion display an embryonic enhancer trap expression pattern (Fig. 4f) very similar to the expression pattern of Cx/Tc-Scr (Curtis et al. 2001; Fig. 4g), despite the fact that the insertion site is about 87 kb upstream of Cx/Tc-Scr. KT076 larvae and pupae also show weak enhancer trap patterns consistent with predicted Cx/Tc-Scr domains (Fig. 4h–i). As expected from the phenotype of homozygotes, KT076 fails to complement ptl D60 as assayed by adult viability, and crosses of KT076 to ptl D60 heterozygotes produce embryos with a phenotype similar to, but slightly weaker than, that of ptl D60 homozygotes (Fig. 4e). In contrast, KT076 fully complements both the embryonic phenotype and the adult viability of Cx 61, a null allele of Cx/Tc-Scr (Shippy et al. 2006), indicating that Cx/Tc-Scr function is not compromised by the insertion (data not shown). These results suggest that KT076 is a hypomorphic allele of ptl/Tc-Antp, and it will, hereafter, be referred to as ptl KT76. Together with the data from the mxp Dch-3 and ptl D60 mutations, the Cx/Tc-Scr-like enhancer trap phenotype of ptl KT76 provides strong evidence that Cx/Tc-Scr regulatory elements are located near, and probably within, ptl/Tc-Antp. Although additional experiments will be necessary to pinpoint the location of regulatory elements and verify this conclusion, it is intriguing to think that overlap of regulatory elements of one Hox gene with the transcription unit of another Hox gene might be an important mechanism of Hox cluster constraint.
Overlap of Cx/Tc-Scr regulatory elements with the ptl/Tc-Antp locus. The gene structure of ptl/Tc-Antp (coding sequence is shaded gray) and the positions of mutant lesions are shown in the diagram. The inferred positions of Cx/Tc-Scr regulatory elements in the ptl/Tc-Antp region are indicated below the diagram
Discussion
The Hox clusters of several insects have now been completely sequenced. While breaks in the cluster seem to have occurred several times in the Drosophila lineage, the clusters of Apis, Anopheles, and Tribolium are intact. This suggests that many insect clusters are still subject to constraints to maintain their organization. Of the insects with intact clusters, Tribolium is, by far, the most genetically tractable and has a strong history of Hox gene studies, thus offering the best system for understanding the constraints acting on an intact insect Hox cluster. Below, we discuss insights provided by our analysis of the Tribolium HOMC into mechanisms that might be responsible for Hox cluster integrity.
Temporal colinearity
Among organisms for which both Hox cluster sequence and expression data are available, the presence of an intact cluster appears to be correlated with temporal colinearity of Hox gene expression, while disrupted clusters are associated with lack of temporal colinearity (Monteiro and Ferrier 2006). This observation has pushed temporal colinearity to the forefront of discussions about Hox cluster maintenance. However, several questions remain to be answered. Does temporal colinearity really require an intact Hox cluster? Is temporal colinearity required for proper Hox gene function in organisms with intact clusters? If temporal colinearity is a key constraint on Tribolium Hox cluster integrity, we might expect rearrangements to affect the function of most or all Hox genes. However, our analysis of Tribolium Hox cluster mutations provides no evidence for such global effects. The ptl D60 inversion splits the complex into two parts but all of the Hox genes except ptl/Tc-Antp and Cx/Tc-Scr appear to function normally. In addition, the mxp Dch-3 rearrangement results in the translocation of several HOMC genes (Tc-zen, Tc-Dfd, Cx/Tc-Scr, and Tc-ftz) to a new chromosomal location. At least one of these genes, Tc-Dfd, is functional because mxp Dch-3 fully complements a Tc-Dfd null allele. Likewise, the genes remaining in the HOMC (with the exception of mxp/Tc-pb) apparently function normally. These limited effects of Hox cluster rearrangements are similar to what has been reported for Drosophila (e.g., Abbott and Kaufman 1986; Pultz et al 1988). Although additional experiments will be required to determine whether Tribolium Hox genes exhibit temporal colinearity, our results suggest that constraints on the Tribolium HOMC are more likely to act locally.
Regulatory elements
The reported breaks and transposition sites in the Hox clusters of Drosophila species are all located in intergenic regions near the 3′ end of a gene (Negre et al. 2005; Negre and Ruiz 2007) and, thus, are presumably less likely to separate a gene from its regulatory elements, which are predominantly located 5′ of each Drosophila Hox gene. Interestingly, the one exception to this rule is the region between abd-A and Abd-B, which contains regulatory regions for both genes and is not split in any of the Drosophila species examined so far (Negre and Ruiz 2007). These observations led to the conclusion that Drosophila Hox clusters have a modular organization (with each gene and its regulatory elements representing a separate module) and that Drosophila Hox genes are still partially clustered simply because the regulatory regions are so large that there are relatively few positions where breaks can occur without disturbing a module. That is, most of the remaining linkage in Drosophila Hox clusters (with the possible exception of abd-A and Abd-B) is due to “phylogenetic inertia,” and, given enough time, the clusters will completely disperse (Negre and Ruiz 2007).
The question naturally arises whether phylogenetic inertia could also be the reason for the intact Hox clusters of insects like Tribolium. Lewis et al. (2003) suggested that unusual features of recombination (Ranz et al. 2001) may make drosophilids more tolerant of Hox cluster rearrangements than are most insects. If this is the case, intact clusters may just be the consequence of slower rates of chromosomal rearrangement (Negre and Ruiz 2007). Alternatively, constraints on Hox cluster maintenance may still be functional in Tribolium.
Our analysis of mutant breakpoints in the ptl/Tc-Antp region indicates that, in at least one case, Tribolium Hox genes are not modular. That is, at least some of the regulatory elements controlling Cx/Tc-Scr expression are apparently located within the pt/Tc-Antp gene (Fig. 5). Most embryonic enhancers of Cx/Tc-Scr are predicted to lie between the mxp Dch-3 and ptl D60 breakpoints because the mxp Dch-3 allele appears to lack all Cx/Tc-Scr function, while ptl D60 has almost normal embryonic Cx/Tc-Scr function. This conclusion is further supported by the expression of mxp/Tc-pb in a Cx/Tc-Scr-like pattern in mxp Dch-3 mutants, presumably due to juxtaposition of the mxp/Tc-pb transcription unit with sequence between Tc-ftz and ptl/Tc-Antp. Adult Cx/Tc-Scr regulatory elements are likely located within, or 5′ of, ptl/Tc-Antp because the ptl D60 rearrangement seems to have a stronger effect on the adult functions of Cx/Tc-Scr. The presence of Cx/Tc-Scr regulatory elements near, or within, ptl/Tc-Antp is supported by the observation that a piggyBac insertion within the ptl/Tc-Antp transcription unit (ptl K76) shows an enhancer trap phenotype that appears to be driven by Cx/Tc-Scr regulatory elements.
In Drosophila, elements which drive Scr-like expression patterns have been found in the region between ftz and Antp (Gorman and Kaufman 1995). However, these elements are apparently redundant because breakpoints in this region only slightly reduce Scr expression levels within its normal domain. Moreover, a deficiency which removes most, if not all, of the Antp transcription unit has no effect on embryonic Scr expression. Thus, it is possible that the modularity of the Drosophila Hox cluster is a recent innovation resulting from changes in the position of cis-regulatory elements. This newly acquired modularity might have allowed breaks in the Hox cluster in the Drosophila lineage. In contrast, overlap of regulatory elements with neighboring genes might still act as a constraint on the integrity of the Hox cluster in Tribolium.
Noncoding transcripts
Although noncoding transcripts within Hox clusters have been recognized for many years (Cumberledge et al. 1990; Lipshitz et al. 1987; Sanchez-Herrero and Akam 1989), there has recently been renewed interest in these enigmatic RNAs. At least two of these ncRNAs, miR-196 and miR-iab-4, can have homeotic function and attenuate the actions of protein-coding Hox genes (Hornstein et al. 2005; Ronshaugen et al. 2005).
In addition to the miRNAs, numerous long noncoding RNAs have been identified within the Hox clusters of both flies and mammals (Bae et al. 2002; Rinn et al. 2007). ncRNAs are implicated in a vast array of processes, including regulation of transcription, translation, epigenetic control of chromatin, mono-allelic expression, dosage compensation, and silencing (reviewed in Mattick and Makunin 2006). In the Drosophila BXC, these transcripts have been implicated in the control of Hox gene expression, although there is some controversy as to whether they promote (Sanchez-Elsner et al. 2006) or repress (Petruk et al. 2006) Hox gene transcription. Mainguy et al. (2007) found evidence for extensive noncoding transcription in the mammalian Hox clusters and proposed that polycistronic and antisense transcription might play a role in keeping Hox genes clustered. Our transcriptional profiling data demonstrates that Tribolium also shows considerable noncoding transcription in the Hox complex. While the tiling array has provided a revealing snapshot of transcription levels during embryonic development, much additional work will be necessary to characterize the actual transcripts. For example, the high levels of transcription in the ptl/Tc-Antp and Utx/Tc-Ubx regions could be the result of several individual transcripts or one long transcript. Interestingly, dicistronic transcripts spanning the Antp and Ubx orthologs have been reported in crustaceans (Shiga et al. 2006) and centipedes (Brena et al. 2006). If such transcripts have an important function, they might constrain linkage in at least some parts of the Hox cluster.
The data available thus far indicate that noncoding transcripts are prevalent in both intact and broken Hox clusters. However, it is not clear whether these transcripts perform the same functions in all organisms. Perhaps, noncoding RNAs play a different or more critical role in organisms with intact clusters. Future studies in this area are likely to provide important insights into Hox gene function and possibly into Hox cluster conservation.
Conclusions
The results presented here provide a foundation for further studies of the constraints acting on Hox clusters. While it is important to keep in mind that multiple factors may have contributed to the maintenance of Hox clusters during evolution (Kmita and Duboule, 2003), the intact structure of the Tribolium Hox cluster and the suite of tools now available for this insect makes it an ideal candidate for such research.
References
Abbott MK, Kaufman TC (1986) The relationship between the functional complexity and the molecular organization of the Antennapedia locus of Drosophila melanogaster. Genetics 114:919–942
Aboobaker A, Blaxter M (2003) Hox gene evolution in nematodes: novelty conserved. Curr Opin Genet Dev 13:593–598
Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA (2002) Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci U S A 99:16847–16852
Beeman RW (1987) A homoeotic gene cluster in the red flour beetle. Nature 327:247–249
Beeman RW, Stauth DM (1997) Rapid cloning of insect transposon insertion junctions using ‘universal’ PCR. Insect Mol Biol 6:83–88
Beeman RW, Stuart JJ, Haas MS, Denell RE (1989) Genetic analysis of the homeotic gene complex (HOMC) in the beetle Tribolium castaneum. Dev Biol 133:196–209
Beeman RW, Stuart JJ, Brown SJ, Denell RE (1993) Structure and function of the homeotic gene complex (HOMC) in the beetle, Tribolium castaneum. Bioessays 15:439–444
Beeman RW, Stuart JJ, Haas MS, Friesen KS (1996) Chromosome extraction and revision of linkage group 2 in Tribolium castaneum. J Hered 87:224–232
Bennett RL, Brown SJ, Denell RE (1999) Molecular and genetic analysis of the Tribolium Ultrabithorax ortholog, Ultrathorax. Dev Genes Evol 209:608–619
Brena C, Chipman AD, Minelli A, Akam M (2006) Expression of trunk Hox genes in the centipede Strigamia maritime: sense and anti-sense transcripts. Evol Dev 8:252–265
Brown SJ, Henry JK, Black WC IV, Denell RE (1990) Molecular genetic manipulation of the red flour beetle: genome organization and cloning of a ribosomal protein gene. Insect Biochem. 20:185–193
Brown S, DeCamillis M, Gonzalez-Charneco K, Denell M, Beeman R, Nie W, Denell R (2000) Implications of the Tribolium Deformed mutant phenotype for the evolution of Hox gene function. Proc Natl Acad Sci U S A 97:4510–4514
Brown SJ, Fellers JP, Shippy TD, Richardson EA, Maxwell M, Stuart JJ, Denell RE (2002) Sequence of the Tribolium castaneum homeotic complex: the region corresponding to the Drosophila melanogaster antennapedia complex. Genetics 160:1067–1074
Cameron RA, Rowen L, Nesbitt R, Bloom S, Rast JP, Berney K, Arenas-Mena C, Martinez P, Lucas S, Richardson PM, Davidson EH, Peterson KJ, Hood L (2006) Unusual gene order and organization of the sea urchin hox cluster. J Exp Zoolog B Mol Dev Evol 306:45–58
Cumberledge S, Zaratzian A, Sakonju S (1990) Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc Natl Acad Sci U S A 87:3259–3263
Curtis CD, Brisson JA, DeCamillis MA, Shippy TD, Brown SJ, Denell RE (2001) Molecular characterization of Cephalothorax, the Tribolium ortholog of Sex combs reduced. Genesis 30:12–20
Dearden PK, Wilson MJ, Sablan L, Osborne PW, Havler M, McNaughton E, Kimura K, Milshina NV, Hasselmann M, Gempe T, Schioett M, Brown SJ, Elsik CG, Holland PW, Kadowaki T, Beye M (2006) Patterns of conservation and change in honey bee developmental genes. Genome Res 16:1376–1384
Duboule D (2007) The rise and fall of Hox gene clusters. Development 134:2549–2560
Ferrier DE, Holland PW (2002) Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol Phylogenet Evol 24:412–417
Ferrier DE, Minguillon C (2003) Evolution of the Hox/ParaHox gene clusters. Int J Dev Biol 47:605–611
Fried C, Prohaska SJ, Stadler PF (2004) Exclusion of repetitive DNA elements from gnathostome Hox clusters. J Exp Zoolog B Mol Dev Evol 302:165–173
Garcia-Fernandez J (2005) Hox, ParaHox, ProtoHox: facts and guesses. Heredity 94:145–152
Garcia-Fernandez J, Holland PW (1994) Archetypal organization of the amphioxus Hox gene cluster. Nature 370:563–566
Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR (1993) Type I repressors of P element mobility. Genetics 135:81–95
Gorman MJ, Kaufman TC (1995) Genetic analysis of embryonic cis-acting regulatory elements of the Drosophila homeotic gene sex combs reduced. Genetics 140:557–572
Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R et al (2002) The genome sequence of the malaria mosquito Anopheles gambiae. Science 298:129–149
Honey Bee Genome Sequence Consortium (2006) Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–949
Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ (2005) The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature 438:671–674
Hughes CL, Kaufman TC (2000) RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development 127:3683–3694
Jimenez G, Paroush Z, Ish-Horowicz D (1997) Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev 11:3072–3082
Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science 296:916–919
Kmita M, Duboule D (2003) Organizing axes in time and space; 25 years of colinear tinkering. Science 301:331–333
Kohany O, Gentles AJ, Hankus L, Jurka J (2006) Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7:474
Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E (2004) Multiplex detection of RNA expression in Drosophila embryos. Science 305:846
Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM (2007) The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol 17:706–710
Lewis EB, Pfeiffer BD, Mathog DR, Celniker SE (2003) Evolution of the homeobox complex in the diptera. Curr Biol 13:587–588
Lipshitz HD, Peattie DA, Hogness DS (1987) Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev 1:307–322
Lorenzen MD, Kimzey T, Shippy TD, Brown SJ, Denell RE, Beeman RW (2007) piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol Biol 16:265–275
Mainguy G, Koster J, Woltering J, Jansen H, Durston A (2007) Extensive polycistronism and antisense transcription in the Mammalian hox clusters. PLoS ONE 2:e356
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15(Spec No 1):R17–R29
Minguillon C, Gardenyes J, Serra E, Castro LF, Hill-Force A, Holland PW, Amemiya CT, Garcia-Fernandez J (2005) No more than 14: the end of the amphioxus Hox cluster. Int J Biol Sci 1:19–23
Monteiro AS, Ferrier DE (2006) Hox genes are not always Colinear. Int J Biol Sci 2:95–103
Negre B, Ruiz A (2007) HOMC evolution in Drosophila: is there a need for Hox gene clustering. Trends Genet 23:55–59
Negre B, Ranz JM, Casals F, Caceres M, Ruiz A (2003) A new split of the Hox gene complex in Drosophila: relocation and evolution of the gene labial. Mol Biol Evol 20:2042–2054
Negre B, Casillas S, Suzanne M, Sanchez-Herrero E, Akam M, Nefedov M, Barbadilla A, de Jong P, Ruiz A (2005) Conservation of regulatory sequences and gene expression patterns in the disintegrating Drosophila Hox gene complex. Genome Res 15:692–700
Nie W, Stronach B, Panganiban G, Shippy T, Brown S, Denell R (2001) Molecular characterization of Tclabial and the 3ô end of the Tribolium homeotic complex. Dev Genes Evol 211:244–251
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623
Paroush Z, Finley RL Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79:805–815
Pattatucci AM, Otteson DC, Kaufman TC (1991) A functional and structural analysis of the Sex combs reduced locus of Drosophila melanogaster. Genetics 129:423–441
Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A (2006) Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127:1209–1221
Pultz MA, Diederich RJ, Cribbs DL, Kaufman TC (1988) The proboscipedia locus of the Antennapedia Complex: a molecular and genetic analysis. Genes Dev 2:901–920
Ranz JM, Casals F, Ruiz A (2001) How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila. Genome Res 11:230–239
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323
Rogers BT, Peterson MD, Kaufman TC (1997) Evolution of the insect body plan as revealed by the Sex combs reduced expression pattern. Development 124:149–157
Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC (2005) The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev 19:2947–252
Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, Finnerty JR (2007) Pre-Bilaterian Origins of the Hox Cluster and the Hox Code: Evidence from the Sea Anemone, Nematostella vectensis. PLoS ONE 2:e153
Sanchez-Herrero E, Akam M (1989) Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107:321–329
Sanchez-Elsner T, Gou D, Kremmer E, Sauer F (2006) Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science 311:1118–1123
Sarkar G, Turner RT, Bolander ME (1993) Restriction-site PCR: a direct method of unknown sequence retrieval adjacent to a known locus by using universal primers. PCR Methods Appl 2:318–322
Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P, Reinhardt R, Chourrout D (2004) Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431:67–71
Shiga Y, Sagawa K, Takai R, Sakaguchi H, Yamagata H (2006) Transcriptional readthrough of Hox genes Ubx and Antp and their divergent post-transcriptional control during crustacean evolution. Evol Dev 8:407–414
Shippy TD, Brown SJ, Denell RE (1998) Molecular characterization of the Tribolium abdominal-A ortholog and implications for the products of the Drosophila gene. Dev Genes Evol 207:446–452
Shippy TD, Guo J, Brown SJ, Beeman RW, Denell RE (2000) Analysis of maxillopedia expression pattern and larval cuticular phenotype in wild-type and mutant tribolium. Genetics 155:721–731
Shippy TD, Rogers CD, Beeman RW, Brown SJ, Denell RE (2006) The Tribolium castaneum ortholog of Sex combs reduced controls dorsal ridge development. Genetics 174:297–307
Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ (2006) Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16:890–900
Stuart JJ, Brown SJ, Beeman RW, Denell RE (1991) A deficiency of the homeotic complex of the beetle Tribolium. Nature 350:72–74
Stuart JJ, Brown SJ, Beeman RW, Denell RE (1993) The Tribolium homeotic gene Abdominal is homologous to abdominal-A of the Drosophila bithorax complex. Development 117:233–243
Tanzer A, Amemiya CT, Kim CB, Stadler PF (2005) Evolution of microRNAs located within Hox gene clusters. J Exp Zoolog B Mol Dev Evol 304:75–85
Tomoyasu Y, Wheeler SR, Denell RE (2005) Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature 433:643–647
Tribolium Genome Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature (in press)
Von Allmen G, Hogga I, Spierer A, Karch F, Bender W, Gyurkovics H, Lewis E (1996) Splits in fruitfly Hox gene complexes. Nature 380:116
Yasukochi Y, Ashakumary LA, Wu C, Yoshido A, Nohata J, Mita K, Sahara K (2004) Organization of the Hox gene cluster of the silkworm, Bombyx mori: a split of the Hox cluster in a non-Drosophila insect. Dev Genes Evol 214:606–614
Yokomori K, Admon A, Goodrich JA, Chen JL, Tjian R (1993) Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev 7:2235–2245
Acknowledgements
We thank Michelle Coleman, Deane Lehmann, Kathy Leonard, Tatum Kimzey, M. Susan Haas, Sandra Koo, Jessica Neely, and Mandar Deshpande for technical assistance. We are grateful to Yoonseong Park for providing EST sequences and to the GEKU piggyBac mutagenesis project (funded by a United States Department of Agriculture grant to S.J.B., R.W.B. R.E.D., Martin Klingler, and Ernst Wimmer) for the ptl KT76 insertion line. Finally, we thank Yoshinori Tomoyasu, Sherry Miller, Renata Bolognesi, Stephen Richards, and Marek Jindra for helpful discussions and comments on the manuscript. This study was funded, in part, by grants from the National Science Foundation (IOB0321882 to R.E.D., S.J.B., and T.D.S.), the National Institutes of Health (HD29594 to S.J.B, R.E.D. and T.D.S, GM34431 to M.L. and GM72395 to M.R.), and the Terry C. Johnson Center for Basic Cancer Research at Kansas State University.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Roth
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Shippy, T.D., Ronshaugen, M., Cande, J. et al. Analysis of the Tribolium homeotic complex: insights into mechanisms constraining insect Hox clusters. Dev Genes Evol 218, 127–139 (2008). https://doi.org/10.1007/s00427-008-0213-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-008-0213-4