Abstract
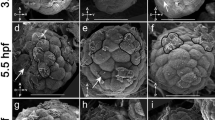
Somitogenesis is the key developmental step, which divides the vertebrate body axis into segmentally repeated structures. It requires an intricate process of pre-patterning, which is driven by an oscillator mechanism consisting of the Delta–Notch pathway and various hairy- and Enhancer of split-related (her) genes. The subset of her genes, which are necessary to set up the segmentation clock, reveal a complex scenario of interactions. To understand which her genes are essential core players in this process, we compared the expression patterns of somitogenesis-relevant her genes in zebrafish and medaka (Oryzias latipes). Most of the respective medaka genes (Ol-her) are duplicated like what has been shown for zebrafish (Dr-her) and pufferfish genes (Fr-her). However, zebrafish genes show some additional copies and significant differences in expression patterns. For the paralogues Dr-her1 and Dr-her11, only one copy exists in the medaka (Ol-her1/11), which combines the expression patterns found for both zebrafish genes. In contrast to Dr-her5, the medaka orthologue appears to play a role in somitogenesis because it is expressed in the presomitic mesoderm (PSM). PSM expression also suggests a role for both Ol-her13 genes, homologues of mouse Hes6 (mHes6), in this process, which would be consistent with a conserved mHes6 homologue gear in the segmentation clock exclusively in lower vertebrates. Members of the mHes5 homologue group seem to be involved in somite formation in all vertebrates (e.g. Dr- and Ol-her12), although different paralogues are additionally recruited in zebrafish (e.g. Dr-her15) and medaka (e.g. Ol-her4). We found that the linkage between duplicates is strongly conserved between pufferfish and medaka and less well conserved in zebrafish. Nevertheless, linkage and orientation of several her duplicates are identical in all three species. Therefore, small-scale duplications must have happened before whole genome duplication occurred in a fish ancestor. Expression of multiple stripes in the intermediate PSM, characteristic for the zebrafish orthologues, is absent in all somitogenesis-related her genes of the medaka. In fact, the expression mode of Ol-her1/11 and Ol-her5 indicates dynamism similar to the hairy clock genes in chicken and mouse. This suggests that Danio rerio shows a rather derived clock mode when compared to other fish species and amniotes or that, alternatively, the clock mode evolved independently in zebrafish, medaka and mouse or chicken.










Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aulehla A, Herrmann BG (2004) Segmentation in vertebrates: clock and gradient finally joined. Genes Dev 18:2060–2067
Bae S-K, Bessho Y, Hojo M, Kageyama R (2000) The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 127:2933–2943
Bessho Y, Kageyama R (2003) Oscillations, clocks and segmentation. Curr Opin Genet Dev 13:379–384
Bessho Y, Miyoshi G, Sakata R, Kageyama R (2001a) Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6:175–185
Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R (2001b) Dynamic expression and essential functions of Hes7 in somite formation. Genes Dev 15:2642–2647
Bessho Y, Hirata H, Masamizu Y, Kageyama R (2003) Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev 17:1451–1456
Cooke J, Zeeman EC (1976) A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol 58:455–476
Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH (2002) Hes6 regulates myogenic differentiation. Development 129:2195–2207
del Barco Barrantes I, Elia AJ, Wünsch K, Hrabe De Angelis M, Mak TW, Rossant J, Conlon RA, Gossler A, de la Pompa JL (1999) Interaction between Notch signaling and Lunatic fringe during somite boundary formation in the mouse. Curr Biol 9:470–480
Draper BW, Stock DW, Kimmel CB (2003) Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development 130:4639–4654
Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37–46
Dubrulle J, Pourquié O (2004) Coupling segmentation to axis formation. Development 131:5783–5793
Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RSP (2002) Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129:1795–1806
Elmasri H, Winkler C, Liedtke D, Sasado T, Morinaga C, Suwa H, Niwa K, Henrich T, Hirose Y, Yasuoka A, Yoda H, Watanabe T, Deguchi T, Iwanami N, Kunimatsu S, Osakada M, Loosli F, Quiring R, Carl M, Grabher C, Winkler S, Del Bene F, Wittbrodt J, Abe K, Takahama Y, Takahashi K, Katada T, Nishina H, Kondoh H, Furutani-Seiki M (2004a) Mutations affecting somite formation in the Medaka (Oryzias latipes). Mech Dev 121:659–671
Elmasri H, Liedtke D, Lücking G, Volff J-N, Gessler M, Winkler C (2004b) her7 and hey1, but not lunatic fringe show dynamic expression during somitogenesis in Medaka (Oryzias latipes). Gene Expr Patterns 4:553–559
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Furutani-Seiki M, Wittbrodt J (2004) Medaka and zebrafish, an evolutionary twin study. Mech Dev 121:629–637
Furutani-Seiki M, Sasado T, Morinaga C, Suwa H, Niwa K, Yoda H, Deguchi T, Hirose Y, Yasuoka A, Henrich T, Watanabe T, Iwanami N, Kitagawa D, Saito K, Asaka S, Osakada M, Kunimatsu S, Elmasri H, Winkler C, Ramialison M, Loosli F, Quiring R, Carl M, Grabher C, Winkler S, Del Bene F, Shinomiya A, Kota Y, Yamanaka T, Okamot Y, Takahashi K, Todo T, Abe K, Takahama Y, Tanaka M, Mitani H, Katada T, Nishina H, Nakajima N, Wittbrodt J, Kondoh H (2004) A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes. Mech Dev 121:647–658
Gajewski M, Voolstra C (2002) Comparative analysis of somitogenesis related genes of the hairy/Enhancer of split class in Fugu and zebrafish. BMC Genomics 3:21
Gajewski M, Sieger D, Alt B, Leve C, Hans S, Wolff C, Rohr KB, Tautz D (2003) Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130:4269–4278
Giudicelli F, Lewis J (2004) The vertebrate segmentation clock. Curr Opin Genet Dev 14:407–414
Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nüsslein-Volhard C (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1–36
Henry CA, Urban MK, Dill KK, Merlie JP, Page MF, Kimmel CB, Amacher SL (2002) Zebrafish her1 and her7 function together to refine alternating somite boundaries. Development 129:3693–3704
Holley SA, Takeda H (2002) Catching a wave: the oscillator and wavefront that create the zebrafish somite. Semin Cell Dev Biol 13:481–488
Holley SA, Geisler R, Nüsslein-Volhard C (2000) Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave front activity. Genes Dev 14:1678–1690
Holley SA, Jülich D, Rauch G-J, Geisle R, Nüsslein-Volhard C (2002) her1 and the Notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129:1175–1183
Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB (2003) Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell 4:67–82
Iwamatsu T (2004) Stages of normal development in the Medaka Oryzias latipes. Mech Dev 121:605–618
Jouve C, Palmeirim I, Henrique D, Beckers J, Gossler A, Ish-Horowicz D, Pourquié O (2000) Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127:1421–1429
Jowett T, Yan YL (1996) Double fluorescence in situ hybridisation to zebrafish embryos. Trends Genet 12:387–389
Jülich D, Hwee Lim C, Round J, Nicolaije C, Schroeder J, Davies A, Geisler R, Lewis J, Jiang YJ, Holley SA, Tübingen 2000 Screen Consortium (2005) beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol 286:391–404
Kawakami Y, Raya A, Raya RM, Rodríguez-Esteban C, Izpisúa Belmonte JC (2005) Retinoic acid signalling links left–right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature 465:165–171
Kawamura A, Koshida S, Hijikata H, Sakaguchi T, Kondoh H, Takada S (2005) Zebrafish Hairy/Enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev 19:1156–1161
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TH (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310
Koyano-Nakagawa N, Kim J, Anderson D, Kintner C (2000) Hes6 acts in a negative feedback loop with the neurogenins to promote neuronal differentiation. Development 127:4203–4216
Lekven AC, Buckles GR, Kostakis N, Moon RT (2003) Wnt1 and wnt10b function redundantly at the zebrafish midbrain–hindbrain boundary. Dev Biol 254:172–187
Leve C, Gajewski M, Rohr KB, Tautz D (2001) Homologues of c-hairy1 (her9) and lunatic fringe in zebrafish are expressed in the developing central nervous system, but not in the presomitic mesoderm. Dev Genes Evol 211:493–500
Lewis KE, Eisen JS (2001) Hedgehog signaling is required for primary motoneuron induction in zebrafish. Development 128:3485–3495
Loosli F, Staub W, Finger-Baier KC, Ober EA, Verkade H, Wittbrodt J, Baier H (2003) Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep 4:894–899
Müller M, v. Weizsäcker E, Campos-Ortega JA (1996). Expression domains of a zebrafish homolog of the Drosophila pair-rule gene hairy correspond to the primordia of alternating somites. Development 122:2071–2078
Naruse K, Tanaka M, Mita K, Shima A, Postlethwait J, Mitani H (2005) A medaka gene map: the traces of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res 14:820–828
Nicholas KB, Nicholas HB, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW NEWS 4:14
Oates AC, Ho RK (2002) Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta–Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129:2929–2946
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12:357–358
Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O (1997) Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91:639–648
Pasini A, Jiang Y-J, Wilkinson DG (2004) Two zebrafish Notch-dependent hairy/Enhancer-of-split-related genes, her6 and her4, are required to maintain the coordination of cyclic gene expression in the presomitic mesoderm. Development 131:1529–1541
Plickert G, Gajewski M, Gehrke G, Gausepohl H, Schlossherr J, Ibrahim H (1997) Automated in situ detection (AISD) of biomolecules. Dev Genes Evol 207:362–367
Pourquié O (2003) The segmentation clock: converting embryonic time into spatial pattern. Science 301:328–330
Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M (1998) Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain–hindbrain boundary development and somitogenesis. Development 125:2381–2395
Rida PC, Le Minh N, Jiang YJ (2004) A Notch feeling of somite segmentation and beyond. Dev Biol 265:2–22
Saga Y, Takeda H (2001) The making of the somite: molecular events in vertebrate segmentation. Nat Rev 2:835–845
Sawada A, Shinya M, Jiang Y-J, Kawakami A, Kuriowa A, Takeda H (2001) Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128:4873–4880
Sieger D, Tautz D, Gajewski M (2003) The role of Suppressor of Hairless in Notch mediated signalling during zebrafish somitogenesis. Mech Dev 120:1083–1094
Sieger D, Tautz D, Gajewski M (2004) her11 is involved in the somitogenesis clock in zebrafish. Dev Genes Evol 214:393–406
Shankaran SS (2004) her15, a novel gene with oscillating mRNA expression domains and its potential role in zebrafish somitogenesis. Ph.D. thesis, University of Cologne
Takke C, Dornseifer P, v. Weizsacker E, Campos-Ortega JA (1999) her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development 126:1811–1821
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang Y-J, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Allende ML, Weinberg ES, Nüsslein-Volhard C (1996) Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123:153–164
Volff J-N (2005) Genome evolution and biodiversity in teleost fish. Heredity 94:280–294
Weizsäcker E v. (1994) Molekulargenetische Untersuchungen an sechs Zebrafisch-Genen mit Homologie zur Enhancer of split Gen-Familie von Drosophila. Ph.D. thesis, Universität zu Köln
Acknowledgements
The authors wish to thank Diethard Tautz, Julian Lewis and Jean-Nicolas Volff for critically reading the manuscript and Manfred Schartl for fruitful discussions and support. Furthermore, the authors from the Department of Evolutionary Genetics thank Diethard Tautz for his generous and constant support of this research project. Genome data have been provided freely by the National Institute of Genetics and the University of Tokyo for use in this publication only. We wish to thank Eva Schetter for excellent technical assistance and David and Carsten for fish care. We are very grateful to Drs. Makoto-Furutani Seiki and Hisato Kondoh for providing the medaka somite mutants. The authors would also like to thank an unknown reviewer for his/her very helpful comments and constructive criticism. The work was supported by the Deutsche Forschungsgemeinschaft (SFB 572) and by the Fond der chemischen Industrie.
Author information
Authors and Affiliations
Corresponding author
Additional information
"M. Gajewski and M. Girschick contributed equally to this work." Communicated by M. Hammerschmidt
An erratum to this article can be found at http://dx.doi.org/10.1007/s00427-006-0072-9
Rights and permissions
About this article
Cite this article
Gajewski, M., Elmasri, H., Girschick, M. et al. Comparative analysis of her genes during fish somitogenesis suggests a mouse/chick-like mode of oscillation in medaka. Dev Genes Evol 216, 315–332 (2006). https://doi.org/10.1007/s00427-006-0059-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-006-0059-6