Abstract.
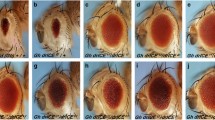
The Drosophila genome contains a single orthologue of mammalian puromycin-sensitive aminopeptidases, dPsa. Even though dPsa was expressed in many tissues during development, animals lacking dPsa activity were viable. Ubiquitous overexpression of dPsa during embryonic or larval development resulted in lethality and overexpression in isolated tissues during development resulted in localized lesions. These results suggest that even though dPsa function was not essential for viability, dPsa expression must be tightly regulated for normal development. By screening the Drosophila genome we found 43 predicted aminopeptidases and generated a phylogenetic tree of aminopeptidases related to dPsa by sequence. We discuss possible functions of dPsa and the idea that other Drosophila aminopeptidases might perform redundant functions with dPsa for regulating protein turnover.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Electronic Publication
Rights and permissions
About this article
Cite this article
Schulz, C., Perezgasga, L. & Fuller, M.T. Genetic analysis of dPsa, the Drosophila orthologue of puromycin-sensitive aminopeptidase, suggests redundancy of aminopeptidases. Dev Genes Evol 211, 581–588 (2001). https://doi.org/10.1007/s00427-001-0194-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-001-0194-z