Abstract
Main conclusions
We have identified new potential regulators of xylem cell-type determination and cellular proliferation in cassava and studied their expression in roots. Results are highly relevant for cassava biotechnology.
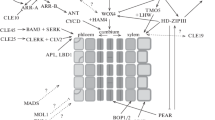
Cassava’s root system is composed of two types of root that coexist in every individual: the fibrous and the storage roots. Whether a root becomes fibrous or storage depends on the xylem cell types that it develops: fibrous roots develop xylem fibres and vessels while storage roots develop parenchyma xylem, the starch-storing tissue. A crucial question in cassava root development is how the specific xylem cell types differentiate and proliferate in the fibrous and storage roots. Using phylogenetic, protein sequence and synteny analyses we identified (1) MeVND6, MeVND7.1, MeVND7.2, MeNST3.1 and MeNST3.2 as the potential cassava orthologues of the Arabidopsis regulators of xylem cell type determination AtVND6, AtVND7 and AtNST3; and (2) MeWOX4.1 and MeWOX4.2 as the potential cassava orthologues of the Arabidopsis cambium regulator AtWOX4. Fibrous and storage roots were anatomically characterised and tested for the expression of the identified genes. Results revealed that (1) MeVND7.1 and MeVND7.2 are expressed in the fibrous but not in the storage roots; (2) MeVND6 shows low expression in both root types; (3) MeNST3.1 is not expressed in the fibrous or storage roots, while MeNST3.2 is highly expressed in both root-types and (4) MeWOX4.1 and, to a higher level, MeWOX4.2 are expressed in both the fibrous and storage roots. Results open new avenues for research in cassava root development and for food security-oriented biotechnology programmes.





Similar content being viewed by others
References
Agusti J, Merelo P, Cercos M, Tadeo FR, Talon M (2008) Ethylene-induced differential gene expression during abscission of citrus leaves. J Exp Bot 59(10):2717–2723
Agusti J, Lichtenberger R, Schwarz M, Nehlin L, Greb T (2011) Characterization of transcriptome remodelling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genet 7(2):e1001312. doi:10.1371/journal.pgen.1001312
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Alves AAC (2002) Cassava botany and physiology. In: Hillcocks RJ, Thresh JM (eds) Cassava: biology, production and utilization. CABI, Wallingford, pp 67–89
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Buschmann H, Potter UJ, Beeching JR (2002) Ultrastructure of cassava root by TEM and SEM. Microsc Anal 87(1):9–11
Ceballos H, Kulakow P, Hershey C (2012) Cassava breeding: current status, bottlenecks and the potential of biotechnology tools. Trop Plant Biol 5(1):73–87
Chaweewan Y, Taylor N (2015) Anatomical assessment of root formation and tuberization in cassava (Manihot esculenta Crantz). Trop Plant Biol 8(1):1–8
El-Sharkawy MA (2004) Cassava biology and physiology. Plant Mol Biol 56(4):481–501
Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10(12):248
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biol 59(3):307–321
Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22(8):2618–2629
Hussey SG, Saidi MN, Hefer CA, Myburg AA, Grima-Pettenati J (2015) Structural, evolutionary and functional analysis of the NAC domain protein family in Eucalyptus. New Phytol 206(4):1337–1350
Ji J, Strable J, Shimizu R, Koenig D, Sinha N, Scanlon MJ (2010) WOX4 promotes procambial development. Plant Physiol 152:1346–1356
Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. doi:10.1093/bioinformatics/btu031
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Gene Dev 19(16):1855–1860
Lebot V (2009) Tropical root and tuber crops. Cassava, sweet potato, yams and aroids. Crop production science in horticulture series (17). CAB books, CABI, Wallingford, UK
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickening and are required for anther dehiscence. Plant Cell 17(11):2993–3006
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19(1):270–280
Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 5(6):288
Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22(10):3461–3473
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10(2):79–87
Prochnik S, Marri PR, Desany B, Rabinowicz PD, Kodira C, Mohiuddin M, Rodriguez F, Fauquet C, Tohme J, Harkins T, Rokhsar DS, Rounsley S (2012) The cassava genome: Current progress, future directions. Trop Plant Biol 5(1):88–94
Reilly K, Bernal D, Cortes DF, Gomez-Vasquez R, Tohme J, Beeching JR (2007) Towards identifying the full set of genes expressed during cassava post-harvest physiological deterioration. Plant Mol Biol 64(1):187–203
Saithong T, Rongsirikul O, Kalapanulak S, Chiewchankaset P, Siriwat W, Netrpan S, Suksangpanomrung M, Meechai A, Cheevadhanarak S (2013) Starch biosynthesis in cassava: a genome-based pathway reconstruction and its exploitation in data integration. BMC Syst Biol 7:75
Sayre R, Beeching JR, Cahoon EB, Egesi C, Fauquet C, Fellman J, Fregene M, Gruissem W, Mallowa S, Manary M, Maziya-Dixon B, Mbanaso A, Schachtman DP, Siritunga D, Taylor N, Vanderschuren H, Zhang P (2011) The BioCassava Plus Program: biofortification of cassava for sub-Saharan Africa. Annu Rev Plant Biol 62:251–272
Schuetz M, Smith R, Ellis B (2013) Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot 64(1):11–31
Sehr EM, Agusti J, Lehner R, Farmer EE, Schwarz M, Greb T (2010) Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signaling in cambium formation. Plant J 63(5):811–822
Sojikul P, Saithong T, Kalapanulak S, Pisuttinusart N, Limsirichaikul S, Tanaka M, Utsimi Y, Sakurai T, Seki M, Narangajavana J (2015) Genome-wide analysis reveals phytohormone action during cassava storage root initiation. Plant Mol Biol 88(6):531–543
Suer S, Agusti J, Sanchez P, Schwarz M, Greb T (2011) WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23(9):3247–3259
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35(Web Server):W71–W74
Whankaew S, Sraphet S, Thaikert R, Boonseng O, Smith DR, Triwitayakorn K (2015) Validation of a reference gene for transcript analysis in cassava (Manihot esculenta Crantz) and its application in analysis of linamarase and α-hydroxynitrile lyase expression at different growth stages. Afr J Biotechnol 14(9):745–751
Xu J, Duan X, Yang J, Beeching JR, Zhang P (2013) Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol 161(3):1517–1528
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18(11):3158–3170
Zhong R, Richardson EA, Ye ZH (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary cell wall biosynthesis in Arabidopsis. Plant Cell 19(9):2776–2791
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20(10):2763–2782
Acknowledgements
We acknowledge John Beeching (University of Bath) for providing plants, David Alabadí (IBMCP-CSIC-UPV, Valencia), Miguel Blazquez (IBMCP-CSIC-UPV, Valencia), Liam Dolan (University of Oxford) and Ana Milhinhos (University of Oxford) for critical review of the manuscript and/or scientific discussions. This work was partially supported by The Royal Society—UK (RG15032).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2016_2623_MOESM1_ESM.pdf
Suppl. Fig. S1 MEME alignments of AtNST3, MeNST3.1, MeNST3.2 identify the NAC domain in all three sequences and a domain present in cassava but not-conserved in Arabidopsis (PDF 200 kb)
425_2016_2623_MOESM2_ESM.pdf
Suppl. Fig. S2 MEME alignments of AtWOX4, MeWOX4.1, MeWOX4.2 identify the homeobox domain and the WUS-Box motif in all three sequences (PDF 204 kb)
425_2016_2623_MOESM3_ESM.pdf
Suppl. Fig. S3 Synteny between the genomic region containing VND6 putative orthologues in Manihot esculenta, Linum usitatissimum, Populus trichocarpa, Salix purpurea, Gossypium raimondii, Theobroma cacao and Arabidopsis thaliana. Each box represents a gene. Boxes point to the gene orientation. Lines connect genes encoding the same putative orthologue across species. Dark grey central boxes represent the VND6 orthologue in each species. White boxes represent unknown proteins or proteins with no function assigned (PDF 78 kb)
425_2016_2623_MOESM4_ESM.pdf
Suppl. Fig. S4 Synteny between the genomic region containing VND7 putative orthologues in Manihot esculenta, Linum usitatissimum, Populus trichocarpa, Salix purpurea, Gossypium raimondii, Theobroma cacao and Arabidopsis thaliana. Each box represents a gene. Boxes point to the gene orientation. Lines connect genes encoding the same putative orthologue across species. Dark grey central boxes represent the VND7 orthologue in each species. White boxes represent unknown proteins or proteins with no function assigned (PDF 78 kb)
425_2016_2623_MOESM5_ESM.pdf
Suppl. Fig. S5 Synteny between the genomic region containing NST3 putative orthologues in Manihot esculenta, Linum usitatissimum, Populus trichocarpa, Gossypium raimondii, Theobroma cacao and Arabidopsis thaliana. Each box represents a gene. Boxes point to the gene orientation. Lines connect genes encoding the same putative orthologue across species. Dark grey central boxes represent the NST3 orthologue in each accession. White boxes represent unknown proteins or proteins with no function assigned (PDF 85 kb)
425_2016_2623_MOESM6_ESM.pdf
Suppl. Fig. S6 Synteny between the genomic region containing WOX4 putative orthologues in Manihot esculenta, Linum usitatissimum, Populus trichocarpa, Salix purpurea, Gossypium raimondii, Theobroma cacao and Arabidopsis thaliana. Each box represents a gene. Boxes point to the gene orientation. Lines connect genes encoding the same putative orthologue across species. Dark grey central boxes represent the WOX4 orthologue in each species. White boxes represent unknown proteins or proteins with no function assigned (PDF 82 kb)
Rights and permissions
About this article
Cite this article
Siebers, T., Catarino, B. & Agusti, J. Identification and expression analyses of new potential regulators of xylem development and cambium activity in cassava (Manihot esculenta). Planta 245, 539–548 (2017). https://doi.org/10.1007/s00425-016-2623-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2623-2