Abstract
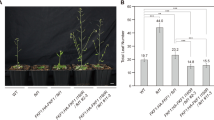
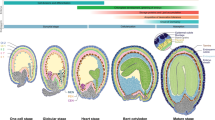
FLOWERING LOCUS C (FLC) is a central floral repressor for the determination of flowering time in Arabidopsis. FLC expression is reactivated upon fertilization and regulated during seed development to ensure the appropriate floral behavior; however, the molecular mechanism for this process is largely unknown. Here, we report the identification of crucial regulators for FLC reactivation during embryogenesis by analyzing FLC::GUS and endogenous FLC expression. We newly define that the full reactivation of FLC requires a FRIGIDA (FRI)-containing protein complex throughout embryogenesis. Mutations in EARLY FLOWERING 7 (ELF7) and VERNALIZATION INDEPENDENCE4 (VIP4) showed severe defects in the reactivation of FLC transcription, suggesting that both of the genes, Arabidopsis homologs of the members of the yeast RNA polymerase II-associated factor 1 (Paf1) complex, are indispensable for FLC reactivation. actin-related protein 6 (arp6), arabidopsis trithorax 1 (atx1), arabidopsis trithorax-related 7 (atxr7), and atx1 atxr7 double mutants also caused the downregulation of FLC during seed development, but the defects were less severe than those in mutants for the FRI- and Paf1-complexes. These results suggest that the ARP6-containing Swr1-complex and FLC-specific histone methyltransferases, ATX1 and ATXR7, have relatively partial roles in FLC reactivation. In contrast to the roles of the histone modifiers, factors in the DNA methylation pathway and biogenesis of small RNAs are not involved in FLC regulation during reproduction. Taken together, our results demonstrate that adjustment by select FLC activators is critical for the re-establishment of an FLC expression state after fertilization to ensure competence for optimal flowering in the next generation.






Similar content being viewed by others
Abbreviations
- Paf1:
-
RNA polymerase II-associated factor 1
- H3K4me3:
-
Trimethylation at histone H3 Lysine4
- DAG:
-
Days after germination
- qRT-PCR:
-
Quatitative RT-PCR
- siRNA:
-
Short interfering RNA
- miRNA:
-
Micro RNA
- RdDM:
-
RNA dependent DNA Methylation
References
Amasino R (2010) Seasonal and developmental timing of flowering. Plant J 61:1001–1013
Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154:516–520
Aufsatz W, Stoiber T, Rakic B, Naumann K (2007) Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene 26:5477–5488
Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36:162–166
Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29:4319–4333
Belotserkovskaya R, Reinberg D (2004) Facts about FACT and transcript elongation through chromatin. Curr Opin Genet Dev 14:139–146
Bender J (2004) DNA methylation and epigenetics. Annu Rev Plant Biol 55:41–68
Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360
Choi J, Hyun Y, Kang MJ, In Yun H, Yun JY, Lister C, Dean C, Amasino RM, Noh B, Noh YS, Choi Y (2009) Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J 57:918–931
Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23:289–303
Choi K, Kim S, Kim SY, Kim M, Hyun Y, Lee H, Choe S, Kim SG, Michaels S, Lee I (2005) SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17:2647–2660
Choi K, Park C, Lee J, Oh M, Noh B, Lee I (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134:1931–1941
Deal RB, Kandasamy MK, McKinney EC, Meagher RB (2005) The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17:2633–2646
Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19:74–83
Dennis ES, Peacock WJ (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10:520–527
Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330:622–627
Finnegan EJ, Genger RK, Kovac K, Peacock WJ, Dennis ES (1998) DNA methylation and the promotion of flowering by vernalization. Proc Natl Acad Sci USA 95:5824–5829
Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107:525–535
Genger RK, Peacock WJ, Dennis ES, Finnegan EJ (2003) Opposing effects of reduced DNA methylation on flowering time in Arabidopsis thaliana. Planta 216:461–466
Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA (2008) Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452:877–881
He Y, Doyle MR, Amasino RM (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18:2774–2784
Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17:1032–1037
Ingouff M, Rademacher S, Holec S, Soljic L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010) Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20:2137–2143
Jean Finnegan E, Kovac KA, Jaligot E, Sheldon CC, James Peacock W, Dennis ES (2005) The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J 44:420–432
Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22:94–97
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347
Kim DH, Doyle MR, Sung S, Amasino RM (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25:277–299
Kim S, Choi K, Park C, Hwang HJ, Lee I (2006) SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18:2985–2998
Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17:3301–3310
Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6:75–83
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16:731–740
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
March-Diaz R, Garcia-Dominguez M, Florencio FJ, Reyes JC (2007) SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol 143:893–901
Martin-Trillo M, Lazaro A, Poethig RS, Gomez-Mena C, Pineiro MA, Martinez-Zapater JM, Jarillo JA (2006) EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133:1241–1252
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13:935–941
Michaels SD, Bezerra IC, Amasino RM (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101:3281–3285
Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137:149–156
Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15:1671–1682
Oh S, Zhang H, Ludwig P, van Nocker S (2004) A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16:2940–2953
Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, Avramova Z, Dean C, Grossniklaus U (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20:580–588
Schlappi MR (2006) FRIGIDA LIKE 2 is a functional allele in Landsberg erecta and compensates for a nonsense allele of FRIGIDA LIKE 1. Plant Physiol 142:1728–1738
Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, Peacock WJ (2008) Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci USA 105:2214–2219
Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97:3753–3758
Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138:1163–1173
Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427:159–164
Swiezewski S, Crevillen P, Liu F, Ecker JR, Jerzmanowski A, Dean C (2007) Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci USA 104:3633–3638
Tamada Y, Yun JY, Woo SC, Amasino RM (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21:3257–3269
Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170:1197–1207
Wu K, Zhang L, Zhou C, Yu CW, Chaikam V (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59:225–234
Xu L, Shen WH (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18:1966–1971
Zhang H, van Nocker S (2002) The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J 31:663–673
Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7:1256–1260
Acknowledgments
We are grateful to S. Michaels for providing the FLC::GUS seeds. We thank R. Amasino, R. Fischer, E. Richard, J. Carrington and Arabidopsis Biological Resource Center (ABRC) for providing mutant seeds. This work was supported by grants from Brain Korea 21 program to H. Yun and Y. Hyun, and from the Korea Research Foundation (KRF-2008-314-C00359) to Y. Choi. This work was also supported by National Research Fund Grant 2009-0079227 from the Ministry of Education, Science and Technology Mid-Career Researcher Program Y. Choi.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yun, H., Hyun, Y., Kang, MJ. et al. Identification of regulators required for the reactivation of FLOWERING LOCUS C during Arabidopsis reproduction. Planta 234, 1237–1250 (2011). https://doi.org/10.1007/s00425-011-1484-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1484-y