Abstract
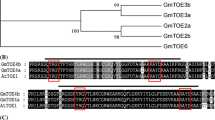
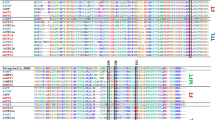
A small gene family of phosphatidyl ethanolamine-binding proteins (PEBP) has been shown to function as key regulators in flowering; in Arabidopsis thaliana the FT protein promotes flowering whilst the closely related TFL1 protein represses flowering. Control of flowering time in soybean [Glycine max (L.) Merrill] is important for geographic adaptation and maximizing yield. Soybean breeders have identified a series of loci, the E-genes, that control photoperiod-mediated flowering time, yet how these loci control flowering is poorly understood. The objectives of this study were to evaluate the expression of GmFT-like genes in the E1 near-isogenic line (NIL) background. Of the 20 closely related PEBP proteins in the soybean genome, ten are similar to the Arabidopsis FT protein. Expression analysis of these ten GmFT-like genes confirmed that only two are detectable in the conditions tested. Further analysis of these two genes in the E1 NILs grown under short-day (SD) and long-day (LD) conditions showed a diurnal expression and tissue specificity expression commensurate with soybean flowering time under SD and LD conditions, suggesting that these were good candidates for flowering induction in soybean. Arabidopsis ft mutant lines flowered early when transformed with the two soybean genes, suggesting that the soybean genes can complement the Arabidopsis FT function. Flowering time in E1 NILs is consistent with the differential expression of the two GmFT-like genes under SD and LD conditions, suggesting that the E1 locus, at least in part, impacts time to flowering through the regulation of soybean FT expression.





Similar content being viewed by others
Abbreviations
- bZIP:
-
Basic region-leucine zipper
- C t :
-
Cycle threshold
- FD:
-
FLOWERING LOCUS D
- FT:
-
FLOWERING LOCUS T
- Gm:
-
Glycine max
- LD:
-
Long day
- NILs:
-
Near isogenic lines
- PEBP:
-
Phosphatidyl ethanolamine-binding protein
- PHYA:
-
PHYTOCHROME A
- qRT-PCR:
-
Real time quantitative reverse transcription PCR
- SD:
-
Short day
- TFL1:
-
TERMINAL FLOWER 1
References
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25:605–614
Bernard RL (1971) Two major genes for time of flowering and maturity in soybeans. Crop Sci 11:242–247
Bonato E, Vello N (1999) E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet Mol Biol 22:229–232
Buzzell RI (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Genet Cytol 13:703
Buzzell RI, Voldeng HD (1980) Inheritance of insensitivity to long daylength. Soybean Genet Newslett 7:26–29
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cober ER, Voldeng HD (2001) Low R:FR light quality delays flowering of E7E7 soybean lines. Crop Sci 41:1823–1926
Cober ER, Tanner JW, Voldeng HD (1996) Genetic control of photoperiod response in early-maturing, near-isogenic soybean lines. Crop Sci 36:601–605
Cober ER, Molnar SJ, Charette M, Voldeng HD (2010) A new locus for early maturity in soybean. Crop Sci 50:524–527
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Faure S, Higgins J, Turner A, Laurie DA (2007) The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176:599–609
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102:7748–7753
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722
Hayama R, Agashe B, Luley E, King R, Coupland G (2007) A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19:2988–3000
Hsu CY, Liu Y, Luthe DS, Yuceer C (2006) Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18:1846–1861
Igasaki T, Watanabe Y, Nishiguchi M, Kotoda N (2008) The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in lombardy poplar. Plant Cell Physiol 49:291–300
Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16:2006–2020
Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6:113–120
Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17:1050–1054
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286:1962–1965
Kobayashi Y, Weigel D (2007) Move on up, it’s time for change–mobile signals controlling photoperiod-dependent flowering. Genes Dev 21:2371–2384
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135:767–774
Kong F-J, Liu B, Xia Z, Sato S, Kim B, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, Abe J (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154:1220–1231
Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103:6398–6403
Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007
Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J (2010) The soybean stem growth habit gene Dt1 Is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153:198–210
Mathieu J, Warthmann N, Kuttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17:1055–1060
McBlain BA, Bernard RL (1987) A new gene affecting the time of flowering and maturity in soybeans. J Hered 178:68–70
Molnar SJ, Rai C, Charette C, Cober ER (2003) Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome 46:1024–1036
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15:473–497
Osborn TC, Kole C, Parkin IA, Sharpe AG, Kuiper M, Lydiate DJ, Trick M (1997) Comparison of flowering time genes in Brassica rapa, B napus and Arabidopsis thaliana. Genetics 146:1123–1129
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang X-C, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci USA 106:8392–8397
Stewart DW, Cober ER, Bernard RL (2003) Modeling genetic effects on the photothermal response of soybean phenological development. Agron J 95:65–70
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Tasma IM, Shoemaker RC (2003) Mapping flowering time gene homologs in soybean and their association with maturity (E) loci. Crop Sci 43:319–328
Thakare D, Kumudini S, Dinkins R (2010) Expression of flowering-time genes in soybean E1 near-isogenic lines under short and long day conditions. Planta 231:951–963
Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Nat Acad Sci USA 107:8563–8568
Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100:13099–13104
Tsuji H, Tamaki S, Komiya R, Shimamoto K (2008) Florigen and the photoperiodic control of flowering in rice. Rice 1:25–35
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59:573–594
Upadhyay AP, Asumadu H, Ellis RH, Qi A (1994a) Characterization of photothermal flowering responses in maturity isolines of soybean [Glycine-max (L) Merrill] Cv. Clark. Annal Bot 74:87–96
Upadhyay AP, Ellis RH, Summerfield RJ, Roberts EH, Qi A (1994b) Variation in the durations of the photoperiod-sensitive and photoperiod-insensitive phases of development to flowering among 8 maturity isolines of soybean [Glycine-max (L) Merrill]. Annal Bot 74:97–101
Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K (2009) Map-based cloning of the gene associated with soybean maturity locus E3. Genetics 182:1251–1262
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Yamagishi N, Yoshikawa N (2011) Expression of FLOWERING LOCUS T from Arabidopsis thaliana induces precocious flowering in soybean irrespective of maturity group and stem growth habit. Planta 233:561–568
Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT. Plant Cell Physiol 46:1175–1189
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Acknowledgments
The authors gratefully acknowledge Dr. Elroy Cober for the E-gene NIL lines, Troy Bass, Elena Prior and Dr. Joseph Omielan for technical assistance, Drs. Karen Hudson and Gary Stacey for manuscript reviews. The use of trade, firm, or corporation names in this paper is for informational purposes only, and does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agriculture Research Service of any product or service to the exclusion of others that may be suitable.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thakare, D., Kumudini, S. & Dinkins, R.D. The alleles at the E1 locus impact the expression pattern of two soybean FT-like genes shown to induce flowering in Arabidopsis. Planta 234, 933–943 (2011). https://doi.org/10.1007/s00425-011-1450-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1450-8