Abstract
We previously showed that recombinant extra domain A from fibronectin (EDA) purified from Escherichia coli was able to bind to toll-like receptor 4 (TLR4) and stimulate production of proinflammatory cytokines by dendritic cells. Because EDA could be used as an adjuvant for vaccine development, we aimed to express it from the tobacco plastome, a promising strategy in molecular farming. To optimize the amount of recombinant EDA (rEDA) in tobacco leaves, different downstream sequences were evaluated as potential fusion tags. Plants generated by tobacco plastid transformation accumulated rEDA at levels up to 2% of the total cellular protein (equivalent to approximately 0.3 mg/g fresh weight) when translationally fused to the first 15 amino acids of green fluorescence protein (GFP). The recombinant adjuvant could be purified from tobacco leaves using a simple procedure, involving ammonium sulfate precipitation and anion exchange chromatography. Purified protein was able to induce production of tumour necrosis factor-α (TNF-α) either by bone marrow-derived dendritic cells or THP-1 monocytes. The rEDA produced in tobacco leaves was also able to induce upregulation of CD54 and CD86 maturation markers on dendritic cells, suggesting that the rEDA retains the proinflammatory properties of the EDA produced in E. coli and thus could be used as an adjuvant in vaccination against infectious agents and cancer. Taken together, these results demonstrate that chloroplasts are an attractive production vehicle for the expression of this protein vaccine adjuvant.
Similar content being viewed by others
Introduction
Transgenic plants are good expression systems for large-scale production of recombinant proteins at industrial levels. Plant systems have many advantages including the low cost of growing plants on a large scale, the availability of natural protein storage organs, and established practices for efficient harvesting, transporting, storing, and processing (Giddings et al. 2000). In the last few years, plants have been genetically engineered to express various recombinant biopharmaceuticals (Ma et al. 2005). However, low production yield is often the most significant limitation in using plants as biofactories (Streatfield 2007). Transgene expression from the plastid genome often allows the very high accumulation of recombinant products, mainly due to high ploidy levels of up to 10,000 plastid genomes per cell (Maliga 2004). Recently, the highest accumulation level ever obtained in a transgenic plant (>70% of the total soluble protein) was reported for tobacco chloroplasts by the expression of a phage-derived protein antibiotic (Oey et al. 2009). Besides high protein yield, plastid transformation offers several advantages, including a much more stable and uniform expression among transgenic lines, the absence of epigenetic effects, the feasibility to express multiple transgenes by linking them together in operons and increased transgene containment due to the maternal inheritance of the plastid genome (reviewed in Maliga 2004). Based on these advantages, several vaccine antigens and biopharmaceuticals have been expressed in tobacco chloroplasts and their efficacy evaluated (Bock 2007; Chebolu and Daniell 2009). For example, vaccine antigens have been expressed against numerous pathogens and shown to be immunogenic and offer protection against the pathogen challenger (Koya et al. 2005; Molina et al. 2005; Arlen et al. 2008). Similarly, several therapeutic human proteins including somatotropin (Staub et al. 2000), interferon alpha (Arlen et al. 2007), proinsulin (Ruhlman et al. 2007) and cardiotrophin-1 (Farran et al. 2008), were expressed in chloroplasts and revealed to be properly folded and fully functional.
Vaccine development remains a high priority for the prevention of diseases for which no vaccine currently exists as well as for improving the efficiency and safety of existing vaccines. Antigen-presenting cells (APCs), especially dendritic cells (DCs), play an important role on the activation of both the innate and adaptive immune responses. Depending on their maturation status, DCs orchestrate a range of immune responses, from tolerance to self-antigens to resistance against infectious pathogens (reviewed in Reis e Sousa 2006). Thus, it is generally accepted that the efficient activation of T cell immune responses is dependent on DC maturation triggered by a combination of stimuli derived from microbial products, endogenous danger signals, or signals delivered by antigen-activated T cells. For these reasons, several vaccination strategies incorporate adjuvant components able to trigger maturation of DCs, either by using ligands for pattern recognition receptors (Cuadros et al. 2004; Lasarte et al. 2007), agonistic antibodies against co-stimulatory molecules (Ahonen et al. 2004) or cytokines (Ahlers et al. 2003).
Some of the most potent DC maturation stimuli are ligands for Toll-like receptors (TLRs) (reviewed in Kaisho and Akira 2002). At present, several ligands for different TLRs have been identified. Most of these ligands are derived from pathogens, although the TLR family is also critical for recognition of certain endogenous molecules. For example, toll-like receptor 4 (TLR4), which is able to recognize lipopolysaccharides (LPS), can also be activated by the spliced exon encoding type III repeat extra domain A from fibronectin (EDA) (Okamura et al. 2001; Lasarte et al. 2007). It has been shown that purified EDA protein produced in E. coli is able to bind to TLR4-expressing cells, activate the TLR4 signaling pathway and stimulate the production of proinflammatory cytokines by DCs and their maturation in vitro and in vivo (Lasarte et al. 2007). Therefore, the EDA protein can be considered as a candidate for vaccine formulations. However, methods for the production of recombinant proteins in high amounts and free of potential contaminants (e.g. bacterial endotoxins) are crucial for vaccine development.
Plants are an attractive system for the production of injectable recombinant proteins, as they do not harbor viruses or prions that are pathogenic in humans. The production of the chicken interferon gamma (IFN-γ) oral vaccine adjuvant, in the endoplasmic reticulum of tobacco plants, has recently been presented (Wu et al. 2009). Despite its high biological activity, which is likely related to its degree of glycosylation, very low expression levels were achieved (up to 0.04% of total soluble protein). IFN-γ has already been produced from the plastid genome of tobacco plants (Leelavathi and Reddy 2003), even if no immunomodulatory activity was reported. In this study, we explored the potential of the plant’s plastid as a bioreactor to produce recombinant EDA (rEDA) as a vaccine adjuvant. A key factor for cost-effective protein production in plastid-transformed plants (transplastomic plants) is the yield of the recombinant protein. Since protein expression in plastids is predominantly controlled at the post-transcriptional level, suitable 5′-untranslated regions (5′-UTRs) are important elements of plastid expression vectors (Farran et al. 2008). Another important feature for high expression of foreign genes in plastids is the sequences immediately downstream of the start codon (Kuroda and Maliga 2001). Modifications or extensions of the N terminus of the desired protein have often led to a remarkable increase in expression levels (Ye et al. 2001; Herz et al. 2005; Lenzi et al. 2008; Zhou et al. 2008; Scotti et al. 2009). Using the well-established tobacco chloroplasts transformation system, we sought to maximize the expression of rEDA by incorporating different fusion tags. The chloroplast-expressed EDA, which retained its proinflammatory properties as an adjuvant, was readily purified from leaves using a simple procedure that did not employ an affinity tag.
Materials and methods
Genetic engineering of the chloroplast expression vectors
The mouse fibronectin EDA sequence was amplified by PCR from the pCR2.1-TOPO-EDA vector (Lasarte et al. 2007) using the upstream primers 5′-TCATGAACATTGATCGCCCTAAAGG-3′ (EDA) and 5′-CCATGGCTAGCATTTCCCCCGGGAACATTGATCGCCCTAAACC-3′ (MEDA), which introduce the BspHI or NcoI site at the start codon, respectively, and the downstream primer 5′-GCGGCCGCTTACTCGAGTGCGGGCGC-3′, which introduces the NotI site. The upstream MEDA primer also encloses a synthetic N-terminal 15-nucleotide fusion tag (Herz et al. 2005) and the SmaI site fused translationally to the 5′-end of EDA. The BspHI-NotI EDA and NcoI-NotI MEDA fragments were fused to the promoter and 5′-UTR of the tobacco psbA gene in a pSK intermediate vector, resulting in plasmids pSK-EDA and pSK-MEDA. Finally, the fusions were digested by EcoRV and NotI and introduced into the tobacco chloroplast transformation vector pAF (Fernandez-San Millan et al. 2008), to produce pAF-EDA and pAF-MEDA.
In addition, three different protein-fusion tags were evaluated. The pSK-GFP:EDA plasmid, which contains the EDA sequence fused at its 5′-end to the first 45 nucleotides of the soluble modified GFP sequence (GENBANK accession no. U_70495), was generated by PCR-based gene assembly using the overlapping primers 5′-TCATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTG-3′ and 5′-CCCGGGAAGAATTGGGACAACTCCAGTGAAAAGTTC-3′ and cloned into the pSK-MEDA plasmid as a BspHI-SmaI fragment. Similarly, pSK-5D1:EDA and pSK-15D1:EDA carry the EDA sequence fused at its 5′-end to the first 15 and 45 nucleotides, respectively, of the tobacco psbA gene which encodes for the D1 protein (GENBANK accession no. NC_001879), were generated by the overlapping primers 5′-TCATGACTGCAATTTTACCCGGG-3′ and 5′-CCCGGGTAAAATTGCAGTCATGA-3′ (5D1:EDA), and 5′-TCATGACTGCAATTTTAGAGAGACGCGAAAGCG-3′ and 5′-CCCGGGACCCCATAGGCTTTCGCTTTCGCGTCTCTC-3′ (15D1:EDA) and cloned into the pSK-MEDA plasmid as a BspHI-SmaI fragment. In all three plasmids, the chimeric genes were expressed from the promoter and 5′UTR of the tobacco psbA gene. The different EDA expression cassettes were introduced into the pAF vector as described above, resulting in pAF-GFP:EDA, pAF-5D1:EDA and pAF-15D1:EDA. The final chloroplast transformation vectors were sequenced and tested for functionality by western-blots of the soluble extracts of E. coli cultures.
Bombardment and regeneration of chloroplast transgenic plants
Gold microprojectiles coated with plasmid DNA (pAF-EDA, pAF-MEDA, pAF-GFP:EDA, pAF-5D1:EDA or pAF-15D1:EDA) were bombarded into in vitro-grown Nicotiana tabacum cv. Petite Havana SR1 (National Germplasm Resources Laboratory, Beltsville, MD, USA) leaves using the PDS1000/He (Bio Rad) biolistic device, as previously described (Daniell 1997). After bombardment of the abaxial leaf side, they were incubated in the dark for 2 days at 28°C. Leaves were then cut into small pieces (~5 × 5 mm) and placed adaxial side up on selection medium (RMOP) in Magenta vessels (Sigma, St. Louis, MO, USA) containing 500 mg/l spectinomycin dihydrochloride as the selecting agent. The regenerated spectinomycin-resistant shoots were subjected to a second round of selection under the same conditions. These regenerated plants were then transplanted and grown in a greenhouse for homoplasmy confirmation and seed production.
Southern-blot and Northern-blot analysis
Total plant DNA (10 µg) was digested with BglII, separated on a 0.8% (w/v) agarose gel and transferred to a nylon membrane. The digestion by BglII and BamHI of the pFS vector generated a 0.8-kb probe (P1) homologous to the flanking sequences. The pSK-MEDA vector digested with SmaI and NotI generated a 0.3-kb probe (P2) homologous to the EDA sequence. Probe labeling and hybridization were performed using the chemiluminiscent AlkPhos direct labeling and detection system (GE Healthcare, Buckinghamshire, UK). After homoplasmy confirmation by Southern analysis, plants were transferred to soil. Seeds from the T0 generation were germinated in vitro on spectinomycin-selective medium. The T1 seedlings were isolated and cultured for 4 weeks in Magenta vessels. Finally, plants were transferred to pots.
Total RNA was extracted (Ultraspec RNA, Biotecx Laboratories) from leaves of transformed and untransformed plants. RNA (10 µg) was separated on 1.5% agarose/formaldehyde gels and then transferred to a nylon membrane. P2 was used as the EDA-specific probe. Labeling and hybridization was performed using the chemiluminiscent detection system mentioned above. Ethidium bromide-stained total leaf RNA was used to asses loading.
Western-blot analyses
Transformed and untransformed leaves of plants grown in a greenhouse were ground in liquid nitrogen. Leaves (100 mg) were homogenized in 200 µl of Laemmli buffer (0.5 M Tris–HCl pH 6.5, 4% SDS, 20% glycerol and 10% β-mercaptoethanol) and heated at 95°C for 5 min. The total cellular protein (TCP) was measured using the RC-DC protein assay (Bio-Rad, Hercules, CA, USA) with BSA as a standard, according to the manufacturer’s instructions. Proteins were separated by SDS–PAGE on 15% polyacrylamide gels and transferred to a nitrocellulose membrane for immunoblotting. Anti EDA-polyclonal antibodies were produced in New Zealand white female rabbits by immunization with 50 µg of EDA mixed with the T helper cell epitope FISEAIIHVLHSR and emulsified in complete Freund’s adjuvant as described by Prieto et al. (1995). Rabbit anti-EDA was used as the primary antibody at a 1:500 dilution and peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma) was used as the secondary antibody at a 1:10,000 dilution. Detection was usually performed using the chemiluminescence ECL western blotting system (GE Healthcare). An enhanced chemiluminescence method (the ECL Advance western blotting detection kit, GE Healthcare) was used when the protein levels were very low. Amounts of rEDA in leaf extracts were determined by comparison with EDA purified from E. coli (3P Biopharmaceuticals, Noain, Spain).
Extraction and purification of recombinant EDA
Fully expanded leaves of pAF-MEDA transformed plants were ground in liquid nitrogen. The ground leaves (~3 g) were homogenized 1:5 (w/v) in protein extraction buffer [20 mM sodium phosphate pH 7.4, 500 mM NaCl, 0.1% (v/v) Triton X-100 and Complete Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany)], and the homogenate was incubated on ice for 30 min. Cell debris were pelleted by centrifugation (20,000×g, 15 min, 4°C) and the supernatant was retained. Ammonium sulfate was added to the supernatant until the salt concentration reached 60% saturation. The mixture was incubated at room temperature for 30 min (first 15 min with agitation) and then centrifuged at 20,000×g for 10 min. The supernatant was retained and mixed with ammonium sulfate until the salt concentration reached 80% saturation. The mixture was incubated and centrifuged as described above. The resulting pellet, resuspended in a 25 mM Tris–HCl pH 7.4 solution, was desalted and concentrated simultaneously by centrifugation (5,000×g, 1 h) in Centricon YM-10 (Millipore, Billerica, MA, USA) centrifugal filter devices (M.W. 10 kDa cut-off). Up to 1 ml of resuspended, desalted and concentrated pellet was loaded onto a Q-Sepharose High Performance 16/100 column (GE Healthcare) equilibrated with a 25 mM Tris–HCl pH 7.4 buffer for anion exchange chromatography with an ÄKTA FPLC system (GE Healthcare). Elution of sample proteins was carried out with a linear gradient of NaCl (0–1 M) for a total of six column volumes at a flow rate of 5 ml/min. Collected fractions were analyzed by western-blot. Fractions containing rEDA were desalted and concentrated by the addition of a 25 mM Tris–HCl pH 7.4 solution, and centrifugation (3,000×g, 40 min) in Centriprep® YM-10 (Millipore) centrifugal filter devices (M.W. 10 kDa cut-off). The protein content of samples was measured with a commercial Bradford assay (Bio-Rad). The purity of rEDA was checked by SDS–PAGE in 15% polyacrylamide gels and Coomassie Blue staining. Before electrophoresis, samples were mixed with loading buffer (0.5 M Tris–HCl pH 6.5, 4% SDS, 20% glycerol, 10% β-mercaptoethanol and 0.1% bromophenol blue) in a 1:1 ratio (1:3 for pellets) and heated at 95°C for 5 min.
The above-described protocol was subsequently adapted to purify the rEDA from the pAF-GFP:EDA plants. Ammonium sulfate at 40% saturation was used in the first step of purification instead of 60%. The rest of the protocol was the same.
In vitro analysis of THP-1 monocyte cell line activation
THP-1 cells (American Type Culture Collection ATCC, Manassas, VA, USA) were used for in vitro assays of monocyte activation by rEDA produced in pAF-MEDA and pAF-GFP:EDA plants. THP-1 cells were plated at 2 × 105 cells/well in 96-well plates and cultured overnight at 37°C and 5% CO2 in complete medium (CM) (RPMI 1640 supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 50 µM 2-mercaptoethanol) (Life Technologies, Cergy-Pontoise, France) for stabilization of the culture. Different concentrations of the indicated antigens were added to the cultures and the culture supernatants were harvested after 15 h of incubation. The concentration of human tumour necrosis factor-α (TNF-α) released to the medium by the THP-1 cell line was quantified using a commercial ELISA assay (BD-Pharmingen, San Diego, CA, USA), according to the manufacturer’s instructions.
In vitro analysis of bone marrow-derived dendritic cell activation
Bone marrow-derived dendritic cells (BMDCs) were generated from mouse femur marrow cell cultures. After lysing erythrocytes with ACK lysis buffer, bone marrow cells were washed and subsequently depleted of lymphocytes and granulocytes by incubation with a mixture of antibodies against CD4 (GK1 clone, ATCC), CD8 (53.6.72 clone, ATCC), Ly-6G/Gr1 (BD-Pharmingen) and CD45R/B220 (BD-Pharmingen) followed by rabbit complement. The remaining cells were grown at 106 cells/ml in 12-well plates in CM supplemented with 20 ng/ml of mGM-CSF and 20 ng/ml of mIL-4 (both from Peprotech, London, UK). Every 2 days, two-thirds of the medium was replaced with fresh medium containing cytokines. Non-adherent dendritic cells were harvested at day 7 and cultured in the presence or absence of different stimuli at 37°C and 5% CO2. After being cultured for 24 h, the supernatants were harvested and TNF-α expression was measured by ELISA (BD-Pharmingen), according to the manufacturer’s instructions.
Expression of DC maturation markers was measured by flow cytometry. Cells were stained with the primary antibodies at 4°C for 15 min, washed and sorted on a FACSscan cytometer (BD Biosciences, San Diego, CA, USA) and analyzed using Cell Quest software (BD Biosciences). The antibodies anti-CD54 (3E2 clone) and anti-CD86 (GL1 clone), both from BD-Pharmingen, were used.
Results
Integration of EDA sequence into the tobacco plastid genome
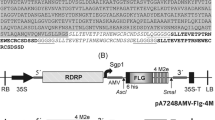
To explore the potential of chloroplasts to produce rEDA, the spliced exon encoding type III repeat extra domain A from fibronectin (Okamura et al. 2001; Lasarte et al. 2007) was expressed from the plastid constitutive promoter of the psbA gene fused to its 5′-UTR, with (MEDA) or without (EDA) an additional 15-nucleotides downstream sequence (DS) which would be expected to add 5 extra-amino acids (MASIS) to the N-terminal of the EDA protein. This sequence has previously been shown to improve protein accumulation (Herz et al. 2005). The chimeric genes were cloned into the chloroplast transformation vector pAF (Fernandez-San Millan et al. 2008) for insertion between the trnI and trnA genes in the tobacco plastome (Fig. 1a). This vector contains a chimeric aadA gene as the selectable marker, which provides spectinomycin resistance for selecting stable transformants.
Integration of chimeric EDA genes into the plastid genome and homoplasmy verification. a Map of the wild-type, EDA and MEDA-transformed plastid genomes. The transgenes are targeted to the intergenic region between trnI and trnA. The EDA/MEDA expression cassette consists of the psbA promoter and 5′-untranslated region (PpsbA) and the psbA terminator (TpsbA) from tobacco. The selectable marker gene aadA is driven by the ribosomal RNA operon promoter (Prrn). The expected sizes of DNA fragments in Southern-blot analysis with the restriction enzyme BglII are indicated. The 0.8-kb fragment (P1) of the targeting region for homologous recombination and the 0.3-kb EDA sequence (P2) were used as probes. Arrows indicate the direction of transcription. M* synthetic N-terminal fusion tag present only in pAF-MEDA construct. Monocistronic transcripts synthesized from the psbA promoter are denoted as asterisk. Di- and polycistronic transcripts, synthesized from the Prrn promoter incorporated with the constructions and read-through transcripts synthesized from the promoter of the rrn operon, are denoted as double and triple asterisk, respectively. The expected sizes of recombinant transcripts are indicated. b, c Southern-blot analysis of two independent transplastomic tobacco lines per construct is shown. Total DNA (10 µg) was digested with BglII and probed with P1 (b) or P2 (c)
The pAF-EDA and pAF-MEDA constructs were introduced into tobacco (cv. Petite Havana) plastids by biolistic transformation (Daniell 1997). Stable integration of foreign genes into the chloroplast genome and homoplasmy of the transformed plants were confirmed by Southern-blot analysis. After two rounds of selection, total leaf DNA from regenerated plants was digested with the BglII restriction enzyme, which cuts in two positions flanking the insertion site and one position in the psbA promoter (Fig. 1a). The 0.8-kb probe P1, which is homologous to the flanking regions trnI and trnA (Fig. 1a), was used. The analyzed transplastomic lines showed the expected hybridization pattern and revealed the absence of residual copies of the wild-type plastome (Fig. 1b). When the blot was stripped and reprobed with the P2 EDA coding region probe (Fig. 1a), hybridization was detected only in the transplastomic lines, as expected (Fig. 1c). To ultimately confirm homoplasmy, seeds from the T0 generation were germinated in vitro on spectinomycin-selective medium. Lack of segregation for spectinomycin resistance in the T1 generation demonstrated homoplasmy (data not shown).
Expression and stability of rEDA in tobacco chloroplasts
The T1 generation pAF-EDA and pAF-MEDA plants were analyzed for levels of rEDA accumulation in leaves. Western-blots of protein extracts from fully expanded leaves of transplastomic plants showed the presence of rEDA only in the pAF-MEDA lines when detected through an anti-EDA polyclonal antibody (Fig. 2a). Leaf extracts of pAF-EDA plants showed no detectable signal (Fig. 2a), even when 50 µg of total cellular protein extracted from young leaves was loaded per lane (data not shown). No immunoreactive polypeptide was present in the wild-type plant extract either, indicating that the signal detected by western-blot was specific for rEDA (Fig. 2a). The expression levels of rEDA in leaves of transplastomic pAF-MEDA plants (MEDA) were determined by comparing the intensity of the immunoreacted bands corresponding to MEDA in the leaf extracts with that of EDA from E. coli. The concentration of MEDA protein was estimated to reach approximately 0.5% of the TCP (Fig. 2b). Monomers of MEDA protein were slightly larger than those produced in E. coli due to the synthetic N-terminal fusion tag used in the pAF-MEDA construct (Fig. 2b). The EDA-expressing transplastomic plants displayed a standard phenotype, indicating that the expression of this protein is phenotypically neutral, at least under standard growth conditions.
Accumulation and stability of rEDA protein in tobacco leaves from pAF-EDA and pAF-MEDA plants. a Western-blot analysis of rEDA in two independent lines per construct. Twenty microgram of total cellular protein were loaded per lane. WT wild-type Petite Havana plant. b Quantification of MEDA protein was determined by analysis of a dilution series of EDA from E. coli (EDA*, 3P Biopharmaceuticals) and leaves extracts from pAF-MEDA plant. The amount of total proteins loaded per lane is indicated. c Northern-blot analysis of total RNA isolated from transplastomic (pAF-EDA and pAF-MEDA lines) and control plants. Ten microgram RNA were electrophoresed in denaturing conditions, blotted onto a Nylon membrane and hybridized with an EDA-specific probe (P2). As load controls, EtBr-stained total leaf RNAs are reported in the lower panel. Mono-, di- and polycistronic transcripts detected by the coding region probe are marked as in Fig. 1a. d rEDA protein accumulation in a developmental series of six alternate leaf samples of a pAF-MEDA plant at the 12-leaf stage (20 µg of total cellular protein each). Leaves are numbered successively from the top (youngest) to the bottom (oldest) of the plant. To assess loading, the Ponceau Red-stained nitrocellulose membrane is also shown. e Northern-blot analysis (as described in c) of a developmental series of leaf samples of a representative pAF-MEDA plant. Leaves were numbered from the top (youngest) to the bottom (oldest)
To study differences in EDA expression, transcript abundance was examined using Northern-blot analysis of total RNA extracted from fully expanded leaves of pAF-EDA and pAF-MEDA lines. Abundant transcripts of the expected size were detected in both lines via hybridization with the P2 specific-EDA probe (Fig. 2c). In all plants, at least three types of transcripts were observed: (1) a monocistronic transcript corresponding to the EDA sequence produced from the psbA promoter; (2) a dicistronic transcript including EDA and aadA sequences produced from the Prrn promoter; and (3) a larger transcript including sequences of EDA, aadA and 16S rRNA arising from read-through transcription from the endogenous promoter of the rrn operon. Consistent with the difference in the coding sequence, transcripts detected in pAF-MEDA plants had slightly lower mobility.
As the chloroplast capacity for protein biosynthesis declines with leaf age, the age-dependent decrease in foreign protein accumulation provides a good indicator of protein stability (Oey et al. 2009). To examine the stability of MEDA in planta, protein extracts from the transplastomic pAF-MEDA plants were prepared from leaves harvested at different positions along the plant. Western-blot analysis showed that MEDA accumulated in all of the analyzed leaves of pAF-MEDA plants (Fig. 2d). The largest amounts of MEDA protein were detected in mature leaves (leaves 5–7; Fig. 2d). A comparison of total RNA isolated from different leaves of the same transplastomic pAF-MEDA plant by Northern-blot showed the presence of specific transcripts in all samples (Fig. 2e), with recombinant mRNA levels even more abundant in older leaves. As a result, rEDA accumulation in tobacco pAF-MEDA plants seemed to be relatively stable along the plant, suggesting that these plants are a good source for protein purification.
Purification of rEDA by ammonium sulfate precipitation and anion exchange chromatography
Partial elimination of contaminating proteins in the leaf extract was achieved by precipitation with ammonium sulfate [(NH4)2SO4]. A range of 40–80% (NH4)2SO4 saturation, using increment steps of 10%, was tested to determine the ammonium sulfate concentration at which the vast majority of MEDA remained soluble while a high proportion of other proteins precipitated. The resulting protein precipitates were analyzed by Coomassie Blue staining and western-blot (Fig. 3a). Because MEDA started to precipitate at 70%, ammonium sulphate at 60% saturation was used in the first step of the purification process, and the plant proteins that precipitated at a lower (NH4)2SO4 concentration were discarded.
Purification steps of rEDA. a Precipitated protein fractions from WT and pAF-MEDA leaf extracts at various ammonium sulfate concentrations. Top panels Coomassie Blue-stained gel of precipitated proteins at different saturating (NH4)2SO4 concentrations (60, 70 and 80%). The pellet was resuspended with three volumes of Laemmli buffer and 10 µl were loaded in each lane. Bottom panels western-blot analysis of the same precipitated ammonium sulfate steps using rabbit anti-EDA as the primary antibody. b Anion exchange chromatography profile of the 60–80% (NH4)2SO4 protein fraction. Chromatograph of rEDA final purification step using a Q-Sepharose High Performance 16/100 column (GE Healthcare) and monitored at 280 nm (thin line). A step gradient of 0–1 M NaCl was applied (thick line) and the fractions were collected. Aliquots of alternate fractions were analyzed by SDS–PAGE and the proteins were detected by western blotting. c Coomassie Blue-stained gel of MEDA purification steps. Purified MEDA is marked on the gel by an arrow. M molecular weight marker, LE leaf extract, P 80 60–80% (NH4)2SO4 protein fraction, FT flow through, P1–4 peaks obtained during gradient elution, EDA* EDA from E. coli (3P Biopharmaceuticals)
Further purification of MEDA was performed by anion exchange chromatography (AEC). The MEDA protein present in the supernatant after the 60% (NH4)2SO4 precipitation step was totally precipitated by an 80% saturated (NH4)2SO4 solution and the dialyzed pellet was loaded onto a Q-Sepharose column. After eluting the sample with a 0–1 M NaCl gradient, western-blot analysis of the obtained fractions showed that MEDA eluted as a single peak (peak 3; Fig. 3b), at a conductivity of ≈33 mS/cm. The identification of AEC peak 3 as the MEDA protein was confirmed by the absence of this peak in the AEC chromatogram of non-transformed Petite Havana control plants subjected to the same purification procedure (data not shown). Analyses by Coomassie Blue staining (Fig. 3c) and silver staining (data not shown) of the eluted peaks also showed that MEDA-containing peak 3 was nearly free of plant contaminating proteins, as these had been almost totally eluted in peaks 1 and 2 (Fig. 3c). The appearance of impurities on the FPLC traces (Fig. 3b, peak 4) but not in the SDS gel (Fig. 3c, lane P4) suggests that these contaminants may not be proteins but other species with an absorbance at 280 nm.
The purity degree of MEDA in each purification step was estimated by densitometry on the Coomassie Blue staining gel (Fig. 3c). In the 60–80% ammonium sulfate protein fraction, the MEDA was approximately 44% pure, whereas the purity reached almost 90% in ACE peak 3.
Recombinant EDA protein produced in tobacco chloroplast induces maturation of bone marrow-derived dendritic cells in vitro
The recombinant fibronectin extra domain A protein produced in E. coli is able to bind to TLR4 and activate the TLR4 signaling pathway and the production of proinflammatory cytokines IL-12 and TNF-alpha by DCs (Lasarte et al. 2007). Accordingly, we wanted to test whether the chloroplast-produced recombinant MEDA would effectively induce activation of both the human monocyte cell line THP-1, which expresses TLR4 molecules (Zhang et al. 1999), and BMDCs in vitro. For this purpose, THP-1 cells or BMDCs were cultured in the presence or absence of rEDA (1 µM) obtained from pAF-MEDA plants, as well as EDA (1 µM) produced in E. coli (3P Biopharmaceuticals), or LPS (0.1 µg/ml) as a positive control. Both rEDA proteins were able to induce the production of the TNF-α cytokine by the human monocyte cell line THP-1 (Fig. 4a). Equivalent results were found when MEDA from three independent production batches was tested (data not shown). Similarly, incubation of BMDCs with 1 µM of MEDA produced in plants, as well as that produced in E. coli, was able to stimulate the production of the TNF-α cytokine (Fig. 4b). Moreover, incubation of BMDCs with 1 µM of EDA protein obtained either from E. coli or pAF-MEDA plants increased the expression of surface molecules CD54 and CD86 (Fig. 4c). These results indicate that recombinant MEDA protein retains the proinflammatory properties previously observed with E. coli-produced EDA.
MEDA protein induced maturation of antigen-presenting cells. a, b Production of proinflammatory cytokine by THP-1 cells (a) or bone marrow-derived dendritic cells (b) incubated for 15 or 24 h, respectively, with or without (negative control) the indicated antigens. Cytokine concentration was measured by ELISA in the culture supernatant. c Flow cytometric analysis of the effects of expression of the co-stimulatory molecules CD54 and CD86 on bone marrow-derived dendritic cells after incubation with the indicated antigens. The data are presented as the mean ± SE of three measurements. EDA* EDA from E. coli (3P Biopharmaceuticals), LPS lipopolysaccharides (positive control), neg negative control, MFI mean fluorescence intensity
Screening of different DS as protein-fusion partners for high yield of rEDA in tobacco chloroplasts
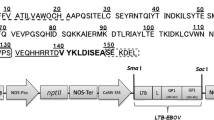
After confirming the proinflammatory properties of the chloroplast-produced EDA protein, we wanted to check the potential for increased rEDA expression in transplastomic tobacco plants. For this purpose, different DS were compared for their improvement of rEDA accumulation in tobacco leaves. Chloroplast transformation was performed with pAF vectors carrying one of three different 5′-tags fused to the EDA protein (Fig. 5a) as a replacement of the additional 15-nucleotide DS employed in the pAF-MEDA construct. As described above, several transplastomic lines were generated from each new construct and purified to homoplasmy. Two representative homoplasmic lines per construct were selected and analyzed for rEDA yield expression by western-blot, and expression levels in fully expanded leaves were compared with those obtained in pAF-MEDA plants (Fig. 5b). Clear differences among the transplastomic lines were observed by western-blot analysis. When the first 5 or 15 amino acids of the photosystem II D1 protein were used as a fusion tag, the amount of recombinant protein measured in those leaves was notably low (Fig. 5b). Thus, the D1 tags caused a drastic decrease of rEDA accumulation in transplastomic tobacco leaves. Moreover, the presence of several abundant shorter immunoreactive bands in the pAF-5D1:EDA and pAF-15D1:EDA plants (Fig. 5b) suggested a high turnover of the recombinant protein in these transplastomic plants. An immunoreactive shorter protein was also detected in protein extracts of pAF-GFP:EDA plants when high levels of recombinant protein were loaded per lane (Figs. 5b, c and 6a). However, relative abundance was lower than for those detected in pAF-D1:EDA plants (Fig. 5b). Interestingly, increased expression levels of rEDA were observed in pAF-GFP:EDA leaf extracts (Fig. 5b). Serial dilutions of EDA from E. coli were used as standards to determine the GFP:EDA concentration by western-blot. The rEDA accumulation in leaves of pAF-GFP:EDA plants was estimated to represent up to 2% of the TCP (Fig. 5c), equivalent to 0.3 mg/g fresh weight. This rEDA protein content represents a fourfold increase in the expression level of that produced in pAF-MEDA plants (Fig. 2b). Northern-blot analysis was performed to determine if changes in EDA mRNA accumulation could account for the remarkable changes in plastid protein accumulation between the different transplastomic lines (Fig. 5d). In all samples, the P2 EDA probe detected specific mono-, di- and polycistronic transcripts to a similar degree, suggesting that the reduction observed in recombinant protein in pAF-D1:EDA plants is mainly due to decreased translation and/or increased protein processing and degradation.
Screening of EDA fusions with different tags and analysis of recombinant protein accumulation in transplastomic plants. a Plastid transformation vectors used in the new transformation experiments. For each vector, the amino acid composition of the downstream sequences fused to the EDA coding N-terminal region is indicated. The two underlined amino acids are generated by the cloning SmaI sequence. The initiating amino acid in the “unmodified” EDA protein is indicated in bold. b Western-blot of proteins extracted from leaves of two independent lines from each construct (pAF-MEDA, pAF-GFP:EDA, pAF-5D1:EDA and pAF-15D1:EDA). Twenty microgram of total cellular protein were loaded per lane. To assess loading, the Ponceau Red-stained nitrocellulose membrane is also shown. WT wild-type Petite Havana plant. c Quantification of rEDA in pAF-GFP:EDA plants was determined by analysis of a dilution series of EDA from E. coli (EDA*, 3P Biopharmaceuticals) and leaves extracts. The amount of total proteins loaded per lane is indicated. d Northern-blot analysis of total RNA isolated from transplastomic (pAF-MEDA, pAF-GFP:EDA, pAF-5D1:EDA and pAF-15D1:EDA lines) and control plants. Ten microgram RNA were electrophoresed in denaturing conditions, blotted onto a Nylon membrane and hybridized with an EDA-specific probe (P2). As load controls, EtBr-stained total leaf RNAs are reported in the lower panel. Mono-, di- and polycistronic transcripts detected by the coding region probe are marked as in Fig. 1a
rEDA protein accumulation and Northern-blot analysis in a developmental series of alternate leaf samples of a representative transplastomic plant of pAF-GFP:EDA, pAF-5D1:EDA and pAF-15D1:EDA lines at the 12-leaf stage. Leaves are numbered successively from the top (youngest) to the bottom (oldest) of the plant. a Western-blot analysis of 20 µg of total cellular protein. Detection in the pAF-5D1:EDA and pAF-15D1:EDA blots was performed using an enhanced chemiluminescence method (ECL Advance western blotting detection kit, GE Healthcare). To assess loading, the Ponceau Red-stained nitrocellulose membrane is also shown. b Total RNA (10 µg per lane) was electrophoresed in denaturing conditions, blotted onto a Nylon membrane and hybridized with an EDA-specific probe (P2). As load controls, EtBr-stained total leaf RNAs are reported in the lower panel. Mono- di- and polycistronic transcripts detected by the coding region probe are marked as in Fig. 1a
To examine rEDA protein accumulation along the plant in pAF-GFP:EDA, pAF-5D1:EDA and pAF-15D1:EDA lines, we investigated its expression in leaves with different development degrees from a mature transplastomic plant (12-leaf stage). Similar to pAF-MEDA, rEDA accumulates in all of the leaves from the pAF-GFP:EDA plants, with the largest protein amounts in mature leaves (Fig. 6a, upper panel). However, the amount of recombinant protein varied with leaf age in pAF-5D1:EDA and pAF-15D1:EDA plants. Protein accumulation was highest in young and fully expanded mature leaves, whereas a general reduction was observed in ageing leaves of both pAF-D1:EDA lines (Fig. 6a, middle and lower panel). Recombinant protein was barely detectable in older leaves of pAF-15D1:EDA plants, even using an enhanced chemiluminescence method of protein detection (Fig. 6a, lower panel). To determine whether these differences in rEDA protein levels were reflected by the RNA-level expression of the transgene, levels of EDA mRNA were examined by probing RNA blots with a specific-EDA probe (Fig. 6b). As demonstrated by the pAF-MEDA plants, mRNA levels in all tested plants slightly increased as leaves aged. As a result, no correlation between transcript accumulation and protein expression level was observed in pAF-D1:EDA plants.
Our results reveal that the fusion protein containing 15 amino acids of GFP resulted in the best yield of rEDA in our studies and could be useful for scaling up protein production in plants by chloroplast transformation. Therefore, an adaptation of the purification protocol described above has been performed to assess the usefulness of rEDA produced in pAF-GFP:EDA plants as adjuvant for vaccination. Considering the similar size and isoelectric point of MEDA and GFP:EDA recombinant proteins, the final purification protocol will not be far from that reported above. Thus, a two-step purification procedure was sufficient to obtain almost pure GFP:EDA protein. An ammonium sulfate cut at 40% saturation was used in the first step of the purification process, followed by anion exchange chromatography. Three independent production batches of recombinant GFP:EDA protein were generated and were able to equivalently induce the production of the TNF-α cytokine by the human monocyte cell line THP-1 (Fig. 7).
Discussion
The development of methods for the production of recombinant proteins in high amounts, free of potential contaminants (i.e., bacterial endotoxins) is of paramount importance for vaccine development. Plants represent an economic and scalable system that could provide modern medicines to developing countries and contribute to biopharmaceutical production in the future. In particular, transgenic chloroplasts offer high levels of foreign protein expression, mainly due to their high transgene polyploidy. Several therapeutic proteins and vaccine antigens have been expressed at high levels via the chloroplast genome and the functionality of chloroplast-derived vaccine antigens and therapeutic proteins has been demonstrated in several assays (reviewed in Chebolu and Daniell 2009). Thus, transgenic chloroplasts are ideal bioreactors for production of functional human and animal therapeutic proteins in an environmentally friendly manner.
We recently demonstrated that the mouse extra domain A from fibronectin might serve as a suitable adjuvant for the development of anti-viral or anti-tumoral vaccines (Lasarte et al. 2007). Here, we report the production of a vaccine adjuvant in tobacco chloroplasts by expressing this protein under the psbA promoter. First, we generated two different transplastomic lines where rEDA was expressed with or without an additional 15-nucleotides DS, which would be expected to add 5 extra-amino acids (MASIS) to the N terminus of the EDA protein. The Northern-blot analysis of transplastomic plants showed that EDA transcripts were correctly transcribed in both transplastomic lines. Nevertheless, recombinant protein accumulation could only be detected in pAF-MEDA plants (up to 0.5% of the TCP), indicating that fusion with the MASIS sequence could partially enhance translation and/or protein stability. The first codons immediately downstream from the AUG have already been identified as an important regulator of translation efficiency in chloroplasts (Kuroda and Maliga 2001). Specifically, the MASIS sequence has been shown to significantly enhance the accumulation of β-glucuronidase (Herz et al. 2005) and HIV-1 p24 antigen (Zhou et al. 2008) in transplastomic tobacco plants. But differential EDA levels could also be explained by differential rates of protein turnover. In this sense, it should be noted that the chimeric EDA gene in pAF-EDA plants codes for Met-Asn-Ile at the N terminus, whereas in pAF-MEDA plants, it codes for Met-Ala-Ser. The putative destabilizing effect of Met-Asn at the N terminus of VP6 (Birch-Machin et al. 2004) and p24 (McCabe et al. 2008) proteins expressed in tobacco chloroplasts has already been described.
Protein turnover during plant development obviously plays an important role in the final accumulation of recombinant proteins. Our data reveal that the accumulation of the rEDA protein in greenhouse-grown transplastomic pAF-MEDA plants is retained in older leaves. The Northern-blot analysis showed a higher accumulation of the recombinant mRNAs in older leaves. By contrast, a decrease of the recombinant mRNA abundance in ageing leaves of transplastomic plants expressing cel6A (Gray et al. 2009) and Pr55gag (Scotti et al. 2009) has been reported. Therefore, the enlarged mRNA levels detected in older leaves of pAF-MEDA plants may be due to an increased transcription rate and/or increased mRNA stability, although stability appears to be the major determinant in our case. In plastids, sequence elements responsible for the regulation of transcript turnover are generally located within the 5′- and 3′-untranslated regions (Eibl et al. 1999). In our constructs, the chimeric genes contain the 5′- and 3′-UTR of the psbA gene. A recent study showed that psbA transcripts accumulated at the highest levels in older leaves of Arabidopsis plants when transcription rates were lowest, consistent with this mRNA’s having high stability (Zoschke et al. 2007).
Our results appear to demonstrate an improvement of rEDA accumulation in tobacco pAF-MEDA plants mediated by the MASIS sequence, although it was not enough to yield high levels of protein accumulation. To overcome this problem, different DS as protein-fusion partners were examined to improve rEDA accumulation in tobacco leaves. The 15-amino acid N-terminal GFP fusion was selected as it was previously shown to increase EPSPS protein accumulation in tobacco chloroplasts (Ye et al. 2001). Our results shown that the pAF-GFP:EDA plants accumulated rEDA up to 2% of the TCP, which represents a fourfold increase of the expression level achieved in pAF-MEDA plants. The equivalent molar quantity (given that the molecular weight of the rEDA = 12.8 kDa) is ~8% of the TCP for a 50 kDa protein, in agreement with the yields reported for the EPSPS protein (47 kDa) when fused to the GFP tag (Ye et al. 2001). Previously, fusion with translation control regions of the plastidial RbcL protein, including both the 5′-UTR and the first 14 amino acids, had increased recombinant protein accumulation in transplastomic tobacco plants (Kuroda and Maliga 2001; Lenzi et al. 2008; Scotti et al. 2009). Because the 5′-UTR of the plastidial psbA gene was used in our constructs, two other DSs based on the first 5 or 15 N-terminal amino acids of the photosystem II D1 protein, which is encoded by the psbA gene, were also evaluated. Moreover, the psbA coding sequence showed evidence of codon bias favoring a maximum translation rate (Morton 1998), so we expected that incorporation of the psbA DS would further enhance rEDA levels. However, we found that plants transformed with these fusion constructs (pAF-5D1:EDA and pAF-15D1:EDA) contained rEDA levels significantly lower than those from plants expressing rEDA from the pAF-MEDA and pAF-GFP:EDA constructs.
Variability in rEDA protein accumulation in these transplastomic plants neither seem to depend on differences at the mRNA level because in Northern-blot analyses, transcripts of the expected size were detected in similar amounts with all vectors. Thus, the N-terminal fusions of EDA protein might affect both the translatability of the mRNA and protein stability. The mechanism of translation enhancement by the DS region remains to be determined, but several DS fusions have been used to increase foreign protein accumulation in tobacco chloroplasts (Ye et al. 2001; Lenzi et al. 2008; Zhou et al. 2008; Scotti et al. 2009). The presence of the 15 amino acids from GFP in pAF-GFP:EDA plants, which could act either by promoting the translation efficiency of transcripts as previously described (Ye et al. 2001) or by increasing the stability of the recombinant protein as recently reported (Molina and Veramendi 2009), might be responsible for the enhanced production of rEDA in the leaves of the transformed plants. However, the increase of rEDA protein accumulation in chloroplasts was greatly lower when DS fusions from D1 protein were used. Recently, a 200-fold increase in E1 catalytic domain expression was reported in transplastomic tobacco plants incorporating the 10 N-terminal amino acids of the D1 protein (Ziegelhoffer et al. 2009), whose increase was attributed to an enhancement in the translation efficiency. Therefore, the low level of rEDA protein detected in pAF-5D1:EDA and pAF-15D1:EDA lines could be gene-specific so that the fusion might create an mRNA structure that interfered with the translation (Kuroda and Maliga 2001). However, the lower accumulation of D1:EDA proteins could also be the result of a decrease in protein stability according to the presence of abundant proteolytic degradation products in western-blots. Thus, the N-terminal D1 tag, with either 5 or 15 amino acids, might in part be responsible for the degradation of the fusion protein. The short half-life of the photosystem II D1 protein (Whitney and Andrews 2001) and the rapid turnover and degradation products detected after the first minutes of synthesis (Kim and Mullet 1994) support this hypothesis. Similar divergent effects on protein yield have also been reported with other fusion tags. For example, very low levels of cellulose Cel6A protein accumulation in transplastomic tobacco plants with the GFP tag were recently described (Gray et al. 2009), which are in contrast with the results for EPSPS (Ye et al. 2001) and EDA (this work). Consequently, in order to enhance protein yields, it is advisable to employ the right fusion partner before proceeding with expression of a transgene in the chloroplast, even though the selection process currently remains highly empirical.
Different patterns of recombinant protein accumulation were detected when expression levels in alternated leaves of these generated transplastomic plants were analysed. As with pAF-MEDA, rEDA accumulates in all of the leaves from the pAF-GFP:EDA plants. However, the amount of D1:EDA proteins declined as the leaves aged, although recombinant transcripts were still present, suggesting that these proteins were more susceptible to proteolytic degradation in chloroplasts than MEDA and GFP:EDA. In fact, a differential stability between the expected size protein and a “shorter than normal” polypeptide could be observed in the oldest leaves of the pAF-15D1:EDA plants, supporting this idea. Similarly, an age-dependent decline in the accumulation of recombinant proteins in tobacco chloroplasts has previously been reported (Birch-Machin et al. 2004; McCabe et al. 2008; Zhou et al. 2008), mainly attributed to increased recombinant protein turnover. Taken together, our results suggest that the additional amino acid sequences encoded by pAF-MEDA and pAF-GFP:EDA not only increase overall protein accumulation but also appear to protect the rEDA proteins from degradation in the older leaves of mature plants. The stability of recombinant proteins throughout the plant is very important for its total yield in transplastomic plants.
The acidic nature (pI 5.2) of human EDA (Miyamoto et al. 2003) allowed for rEDA purification by anion exchange chromatography, similar to other acidic proteins, such as HIV-1 p24 antigen (McCabe et al. 2008), human prothymosin alpha (Yi et al. 2008) and plant lectin (Yan et al. 2005). Moreover, the high stability of the MEDA protein at high ammonium sulfate concentrations allowed for its enrichment before AEC. As such, a two-step purification procedure was sufficient to obtain pure MEDA. The first step of precipitation with 60% saturated (NH4)2SO4 removed 43% of plant contaminating proteins, whereas the second step of AEC eliminated 79% of the remaining protein contaminants. In particular, the second step of AEC resulted in a single peak for MEDA protein with a nearly homogeneous purity, as shown by Coomassie Blue staining. The yield obtained with this protocol was approximately 60–70 mg of ~90% pure rEDA per kg of tobacco leaves, with an estimated recovery of 54%. This protocol is ideal, as purification using affinity tags requires expensive resins that cannot be employed on a large scale. Furthermore, the therapeutic use of plant protein extracts is safer than from bacterial lysates and avoids expensive purification steps to eliminate bacterial endotoxins. Because the rEDA yield could be improved by four times when using pAF-GFP:EDA plants, an adaptation of this purification protocol to the GFP:EDA protein has been performed so that the ammonium sulfate cut was made at 40% saturation instead of 60%. Hence, we should be able to generate 240–280 mg of pure GFP:EDA from 1 kg of tobacco leaves at maturity in the greenhouse. Based on the observed expression levels and considering a potential yield of 50,000 kg/ha of leaves for a commercial tobacco cultivar, it should be possible to obtain yields up to 12–14 kg of pure rEDA in 1 ha of tobacco plants. This yield would be from a single harvest of biomass; however, a single hectare of tobacco biomass can be harvested four to six times within a growing season, depending on the latitude.
Purified rEDA from pAF-MEDA plants was shown to be active in different in vitro assays; it was able to induce production of proinflammatory cytokines by human THP-1 monocytes and bone marrow-derived dendritic cells. The proinflammatory capacity of MEDA obtained from tobacco chloroplasts was slightly lower than the E. coli-produced protein, but very similar to the proinflammatory capacity of LPS, a potent immunomodulator and inducer of inflammatory cytokines. Interestingly, MEDA was also able to induce upregulation of DC maturation markers similar to that by EDA from E. coli or LPS. Likewise, purified GFP:EDA also stimulated the production of TNF-α cytokine by human THP-1 monocytes, demonstrating that the retained extra-amino acids at the N terminus of both MEDA and GFP:EDA proteins do not alter the immunological properties of the EDA protein. These results may have important applications in vaccine development since it has been shown that when DCs fail to mature after taking up the antigen, tolerance rather than immunity may be induced (Steinman et al. 2003).
Considering the potential advantages of plastid expression for biopharming (Bock 2007; Chebolu and Daniell 2009), our results demonstrate the feasibility of using chloroplasts as an ideal expression platform for vaccine adjuvant proteins. In conclusion, we have successfully expressed rEDA protein in tobacco plants at high levels for commercial purposes. We also present an efficient purification protocol using a simple two-step procedure and demonstrate that the resultant pure rEDA retains its proinflammatory properties. Moreover, for oral vaccines, the recombinant adjuvant (EDA) could be co-expressed with the antigen in a transplastomic edible plant avoiding costly purification steps for vaccine formulation.
Abbreviations
- BMDC:
-
Bone marrow-derived dendritic cell
- DC:
-
Dendritic cell
- DS:
-
Downstream sequence
- EDA:
-
Extra domain A from fibronectin
- TCP:
-
Total cellular protein
- TLR:
-
Toll-like receptor
- TNF-α:
-
Tumour necrosis factor-α
References
Ahlers JD, Belyakov IM, Berzofsky JA (2003) Cytokine, chemokine, and costimulatory molecule modulation to enhance efficacy of HIV vaccines. Curr Mol Med 3:285–301
Ahonen CL, Doxsee CL, McGurran SM, Riter TR, Wade WF, Barth RJ, Vasilakos JP, Noelle RJ, Kedl RM (2004) Combined TLR and CD40 triggering induces potent CD8+ T cell expansion with variable dependence on type I IFN. J Exp Med 199:775–784
Arlen PA, Falconer R, Cherukumilli S, Cole A, Cole AM, Oishi KK, Daniell H (2007) Field production and functional evaluation of chloroplast-derived interferon-alpha2b. Plant Biotechnol J 5:511–525
Arlen PA, Singleton M, Adamovicz JJ, Ding Y, Davoodi-Semiromi A, Daniell H (2008) Effective plague vaccination via oral delivery of plant cells expressing F1-V antigens in chloroplasts. Infect Immun 76:3640–3650
Birch-Machin I, Newell CA, Hibberd JM, Gray JC (2004) Accumulation of rotavirus VP6 protein in chloroplasts of transplastomic tobacco is limited by protein stability. Plant Biotechnol J 2:261–270
Bock R (2007) Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol 18:100–106
Chebolu S, Daniell H (2009) Chloroplast-derived vaccine antigens and biopharmaceuticals: expression, folding, assembly and functionality. Curr Top Microbiol Immunol 332:33–54
Cuadros C, Lopez-Hernandez FJ, Dominguez AL, McClelland M, Lustgarten J (2004) Flagellin fusion proteins as adjuvants or vaccines induce specific immune responses. Infect Immun 72:2810–2816
Daniell H (1997) Transformation and foreign gene expression in plants by microprojectile bombardment. Methods Mol Biol 62:463–489
Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop HU (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19:333–345
Farran I, Rio-Manterola F, Iniguez M, Garate S, Prieto J, Mingo-Castel AM (2008) High-density seedling expression system for the production of bioactive human cardiotrophin-1, a potential therapeutic cytokine, in transgenic tobacco chloroplasts. Plant Biotechnol J 6:516–527
Fernandez-San Millan A, Ortigosa SM, Hervas-Stubbs S, Corral-Martinez P, Segui-Simarro JM, Gaetan J, Coursaget P, Veramendi J (2008) Human papillomavirus L1 protein expressed in tobacco chloroplasts self-assembles into virus-like particles that are highly immunogenic. Plant Biotechnol J 6:427–441
Giddings G, Allison G, Brooks D, Carter A (2000) Transgenic plants as factories for biopharmaceuticals. Nat Biotechnol 18:1151–1155
Gray BN, Ahner BA, Hanson MR (2009) High-level bacterial cellulase accumulation in chloroplast-transformed tobacco mediated by downstream box fusions. Biotechnol Bioeng 102:1045–1054
Herz S, Fussl M, Steiger S, Koop HU (2005) Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic Res 14:969–982
Kaisho T, Akira S (2002) Toll-like receptors as adjuvant receptors. Biochim Biophys Acta 1589:1–13
Kim J, Mullet JE (1994) Ribosome-binding sites on chloroplast rbcL and psbA mRNAs and light-induced initiation of D1 translation. Plant Mol Biol 25:437–448
Koya V, Moayeri M, Leppla SH, Daniell H (2005) Plant-based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect Immun 73:8266–8274
Kuroda H, Maliga P (2001) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol 125:430–436
Lasarte JJ, Casares N, Gorraiz M, Hervas-Stubbs S, Arribillaga L, Mansilla C, Durantez M, Llopiz D, Sarobe P, Borras-Cuesta F, Prieto J, Leclerc C (2007) The extra domain A from fibronectin targets antigens to TLR4-expressing cells and induces cytotoxic T cell responses in vivo. J Immunol 178:748–756
Leelavathi S, Reddy VS (2003) Chloroplast expression of His-tagged GUS-fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol Breed 11:49–58
Lenzi P, Scotti N, Alagna F, Tornesello ML, Pompa A, Vitale A, De Stradis A, Monti L, Grillo S, Buonaguro FM, Maliga P, Cardi T (2008) Translational fusion of chloroplast-expressed human papillomavirus type 16 L1 capsid protein enhances antigen accumulation in transplastomic tobacco. Transgenic Res 17:1091–1102
Ma JK, Barros E, Bock R, Christou P, Dale PJ, Dix PJ, Fischer R, Irwin J, Mahoney R, Pezzotti M, Schillberg S, Sparrow P, Stoger E, Twyman RM (2005) Molecular farming for new drugs and vaccines. Current perspectives on the production of pharmaceuticals in transgenic plants. EMBO Rep 6:593–599
Maliga P (2004) Plastid transformation in higher plants. Annu Rev Plant Biol 55:289–313
McCabe MS, Klaas M, Gonzalez-Rabade N, Poage M, Badillo-Corona JA, Zhou F, Karcher D, Bock R, Gray JC, Dix PJ (2008) Plastid transformation of high-biomass tobacco variety Maryland Mammoth for production of human immunodeficiency virus type 1 (HIV-1) p24 antigen. Plant Biotechnol J 6:914–929
Miyamoto K, Kobayashi D, Maeda R, Ito T, Komai T (2003) Inhibition of cryogelation by the novel synthetic peptide (Gly-Arg-Lys-Lys-Thr): recognition site of extra domain A containing fibronectin for heparin in cryogelation. Int J Biol Macromol 31:207–215
Molina A, Veramendi J (2009) High stability of recombinant proteins expressed in tobacco chloroplasts. Open Biotechnol J. pp 67–72
Molina A, Veramendi J, Hervas-Stubbs S (2005) Induction of neutralizing antibodies by a tobacco chloroplast-derived vaccine based on a B cell epitope from canine parvovirus. Virology 342:266–275
Morton BR (1998) Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J Mol Evol 46:449–459
Oey M, Lohse M, Kreikemeyer B, Bock R (2009) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57:436–445
Okamura Y, Watari M, Jerud ES, Young DW, Ishizaka ST, Rose J, Chow JC, Strauss JF 3rd (2001) The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem 276:10229–10233
Prieto I, Hervas-Stubbs S, Garcia-Granero M, Berasain C, Riezu-Boj JI, Lasarte JJ, Sarobe P, Prieto J, Borras-Cuesta F (1995) Simple strategy to induce antibodies of distinct specificity: application to the mapping of gp120 and inhibition of HIV-1 infectivity. Eur J Immunol 25:877–883
Reis e Sousa C (2006) Dendritic cells in a mature age. Nat Rev Immunol 6:476–483
Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H (2007) Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts––oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J 5:495–510
Scotti N, Alagna F, Ferraiolo E, Formisano G, Sannino L, Buonaguro L, De Stradis A, Vitale A, Monti L, Grillo S, Buonaguro FM, Cardi T (2009) High-level expression of the HIV-1 Pr55gag polyprotein in transgenic tobacco chloroplasts. Planta 229:1109–1122
Staub JM, Garcia B, Graves J, Hajdukiewicz PT, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, Ward D, Ye G, Russell DA (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18:333–338
Steinman RM, Granelli-Piperno A, Pope M, Trumpfheller C, Ignatius R, Arrode G, Racz P, Tenner-Racz K (2003) The interaction of immunodeficiency viruses with dendritic cells. Curr Top Microbiol Immunol 276:1–30
Streatfield SJ (2007) Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnol J 5:2–15
Whitney SM, Andrews TJ (2001) The gene for the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13:193–205
Wu Y, Zhao D, Song L, Xu W (2009) Heterologous expression of synthetic chicken IFN-gamma in transgenic tobacco plants. Biologia 64:1115–1122
Yan Q, Jiang Z, Yang S, Deng W, Han L (2005) A novel homodimeric lectin from Astragalus mongholicus with antifungal activity. Arch Biochem Biophys 442:72–81
Ye GN, Hajdukiewicz PT, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM (2001) Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J 25:261–270
Yi S, Brickenden A, Choy WY (2008) A new protocol for high-yield purification of recombinant human prothymosin alpha expressed in Escherichia coli for NMR studies. Protein Expr Purif 57:1–8
Zhang FX, Kirschning CJ, Mancinelli R, Xu XP, Jin Y, Faure E, Mantovani A, Rothe M, Muzio M, Arditi M (1999) Bacterial lipopolysaccharide activates nuclear factor-kappaB through interleukin-1 signaling mediators in cultured human dermal endothelial cells and mononuclear phagocytes. J Biol Chem 274:7611–7614
Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers AM, Maloney AP, Kavanagh TA, Gray JC, Bock R (2008) High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J 6:897–913
Ziegelhoffer T, Raasch JA, Austin-Phillips S (2009) Expression of Acidothermus cellulolyticus E1 endo-beta-1, 4-glucanase catalytic domain in transplastomic tobacco. Plant Biotechnol J 7:527–536
Zoschke R, Liere K, Borner T (2007) From seedling to mature plant: arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50:710–722
Acknowledgments
The authors wish to thank P. Obregon and J. Veramendi, UPNA, Pamplona, Spain for a critical reading of this manuscript. The authors also thank K. Recalde and I. Serrano, UPNA, Pamplona, Spain for technical assistance. This work was supported by a grant (Proyecto EUROINNOVA) from the Department of Innovation (Gobierno de Navarra).
Author information
Authors and Affiliations
Corresponding author
Additional information
I. Farran and I. McCarthy-Suárez contributed equally to this article.
The authors dedicate this publication to the memory of their colleague and co-author, Dr. Ángel M. Mingo-Castel, who died on 7 June 2009.
Rights and permissions
About this article
Cite this article
Farran, I., McCarthy-Suárez, I., Río-Manterola, F. et al. The vaccine adjuvant extra domain A from fibronectin retains its proinflammatory properties when expressed in tobacco chloroplasts. Planta 231, 977–990 (2010). https://doi.org/10.1007/s00425-010-1102-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1102-4