Abstract
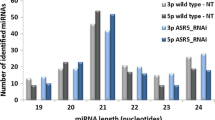
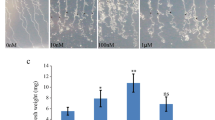
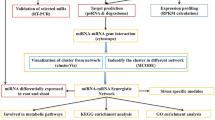
Auxin is one of the central hormones in plants, and auxin response factor (ARF) is a key regulator in the early auxin response. MicroRNAs (miRNAs) play an essential role in auxin signal transduction, but knowledge remains limited about the regulatory network between miRNAs and protein-coding genes (e.g. ARFs) involved in auxin signalling. In this study, we used a novel auxin-resistant rice mutant with plethoric root defects to investigate the miRNA expression patterns using microarray analysis. A number of miRNAs showed reduced auxin sensitivity in the mutant compared with the wild type, consistent with the auxin-resistant phenotype of the mutant. Four miRNAs with significantly altered expression patterns in the mutant were further confirmed by Northern blot, which supported our microarray data. Clustering analysis revealed some novel auxin-sensitive miRNAs in roots. Analysis of miRNA duplication and expression patterns suggested the evolutionary conservation between miRNAs and protein-coding genes. MiRNA promoter analysis suggested the possibility that most plant miRNAs might share the similar transcriptional mechanisms with other non-plant eukaryotic genes transcribed by RNA polymerase II. Auxin response elements were proved to be more frequently present in auxin-related miRNA promoters. Comparative analysis of miRNA and protein-coding gene expression datasets uncovered many reciprocally expressed miRNA–target pairs, which could provide some hints for miRNA downstream analysis. Based on these findings, we also proposed a feedback circuit between miRNA(s) and ARF(s). The results presented here could serve as the basis for further in-depth studies of plant miRNAs involved in auxin signalling.






Similar content being viewed by others
Abbreviations
- α-NAA:
-
α-Naphthalene acetic acid
- ARF:
-
Auxin response factor
- AuxRE:
-
Auxin response element
- EMS:
-
Ethyl methanesulfonate
- GO:
-
Gene Ontology
- IAA:
-
3-Indole acetic acid
- MiRNA:
-
MicroRNA
- MT:
-
osaxr mutant
- osaxr :
-
Oryza sativa auxin resistant
- TF:
-
Transcription factor
- TSS:
-
Transcription start site
- WT:
-
Wild type
References
Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283:15932–15945
Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13:343–349
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Breathnach R, Chambon P (1981) Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383
Chiou TJ (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30:323–332
Cleveland WS, Devlin SJ (1988) Locally weighted regression: an approach to regression analysis by local fitting. J Am Stat Assoc 83:596–610
Cui X, Xu SM, Mu DS, Yang ZM (2009) Genomic analysis of rice microRNA promoters and clusters. Gene 431:61–66
Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9:302–308
Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235–1250
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Gao X, Gulari E, Zhou X (2004) In situ synthesis of oligonucleotide microarrays. Biopolymers 73:579–596
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD (2005) ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res 33:D637–D640
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314
Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17:1387–1396
Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6:47–59
Jiang D, Yin C, Yu A, Zhou X, Liang W, Yuan Z, Xu Y, Yu Q, Wen T, Zhang D (2006) Duplication and expression analysis of multicopy miRNA gene family members in Arabidopsis and rice. Cell Res 16:507–518
Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V (2007) CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res 35:D829–D833
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53:377–398
Li A, Mao L (2007) Evolution of plant microRNA gene families. Cell Res 17:212–218
Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98:31–36
Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52:133–146
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Maher C, Stein L, Ware D (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16:510–519
Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375
Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG (2006) MicroRNA promoter element discovery in Arabidopsis. RNA 12:1612–1619
Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49:592–606
Ru P, Xu L, Ma H, Huang H (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res 16:457–465
Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV (2003) PlantProm: a database of plant promoter sequences. Nucleic Acids Res 31:114–117
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627
Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNAs from rice. Plant Cell 17:1397–1411
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301–309
Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7:847–859
Tian CE, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT (2004) Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J 40:333–343
Tsang J, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26:753–767
Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7:1611–1623
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17:2204–2216
Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA 102:9703–9708
Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133:4211–4218
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Yang JH, Han SJ, Yoon EK, Lee WS (2006) Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res 34:1892–1899
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. The International Rice Research Institute, Manila
Zhang Y (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33:W701–W704
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354:585–590
Zhou X, Ruan J, Wang G, Zhang W (2007) Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 3:e37
Zhu Q, Hong A, Sheng N, Zhang X, Matejko A, Jun KY, Srivannavit O, Gulari E, Gao X, Zhou X (2007) microParaflo biochip for nucleic acid and protein analysis. Methods Mol Biol 382:287–312
Acknowledgments
This study is supported by 973 Program (2005CB120901), NSFC (30771326), 863 Program (2008AA10Z125), and Program for NCET.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
This file is unfortunately not in the Publisher's archive anymore: Supplementary material 10 (RAR 7642 kb)
Rights and permissions
About this article
Cite this article
Meng, Y., Huang, F., Shi, Q. et al. Genome-wide survey of rice microRNAs and microRNA–target pairs in the root of a novel auxin-resistant mutant. Planta 230, 883–898 (2009). https://doi.org/10.1007/s00425-009-0994-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-0994-3