Abstract
Abscisic Acid (ABA) is an important phytohormone involved in abiotic stress resistance in plants. A group of bZIP transcription factors play important roles in the ABA signaling pathway in Arabidopsis. However, little is known about the function of their orthologs in rice, where they may hold a great potential for developing drought resistant food crops. In this study, our phylogenetic analysis showed that this group of bZIPs was evolutionarily conserved between Arabidopsis and rice, which implies that they may share similar functions. We demonstrated with quantitative RT-PCR that the expressions of most of these OsbZIPs were significantly induced by ABA, ACC, and abiotic stresses. OsbZIP72, a member of this group, was proved to be an ABRE binding factor in rice using the yeast hybrid systems. We showed that it could bind to ABRE and transactivate the downstream reporter genes in yeast, and the transactivity was depending on its N-terminal region. Transgenic rice overexpressing OsbZIP72 showed a hypersensitivity to ABA, elevated levels of expression of ABA response gene such as LEAs, and an enhanced ability of drought tolerance. These results suggest that OsbZIP72 plays a positive role in drought resistance through ABA signaling, and is potential useful for engineering drought tolerant rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice that provides staple food for half of the world population relies heavily on water availability for cultivation. As the shortage of fresh water for agricultural irrigation worsens, it becomes increasingly urgent to develop drought resistance crops. The improvement of crop drought resistance through biotechnology holds a great potential for increasing food production with limited fresh water resources, and requires a better understanding of the molecular mechanisms of the drought resistance in plants (e.g., Xiong et al. 2002; Zhu 2002; Wang et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2006; Seki et al. 2007).
Because the phytohormone abscisic acid (ABA) plays an essential role in plant adaption to drought stress, many molecular biology studies have characterized genes responsive to both stress and exogenous ABA. When stress was sensed by plants, the ABA level was elevated, which consequently activated the ABA response genes that trigger physiological reactions for drought resistance. Promoter analysis of the ABA responsive genes revealed that ABA responsive elements (ABREs) harbored in their promoters were major cis-elements for the trans-activation of these genes in response to ABA (Guiltinan et al. 1990; Mundy et al. 1990; Busk and Pages 1998), and a major group of transactivation factors binding to ABREs were basic leucine zipper proteins (bZIPs) that contained basic regions for DNA binding and leucine zipper regions for dimerization (Kim et al. 1997; Uno et al. 2000; Choi et al. 2000).
The bZIPs were best characterized in the model species Arabidopsis thaliana. A total of 75 bZIPs classified into 10 groups have been identified in Arabidopsis (Jakoby et al. 2002). Most of the ABRE binding bZIPs belongs to group A. Functional characterization of several bZIPs in this group revealed that their expression could be triggered by ABA or abiotic stresses (Uno et al. 2000; Choi et al. 2000; Finkelstein and Lynch 2000), and their activations were also regulated by the ABA depended phosphorylation (Lopez-Molina et al. 2001; Furihata et al. 2006). For example, expression level, protein stability, phosphorylation status, and activity of ABI5 were highly correlated with the ABA level, and Arabidopsis overexpressing ABI5 was hypersensitive to ABA during both germination and vegetative growth stages (Lopez-Molina et al. 2001). Transgenic Arabidopsis plants constitutively expressing ABRE binding factor (ABF), ABF3 or ABF4, were also hypersensitive to ABA and showed enhanced drought tolerance (Kang et al. 2002). The overexpression of ABF3 in rice also enhanced drought tolerance (Oh et al. 2005). A rice ABF gene, TRAB1, was found involved in the ABA-regulated transcription (Hobo et al. 1999), and overexpression of the rice orthologs of ABI5 also enhanced ABA sensitivity in Arabidopsis while repression of this gene in rice enhanced abiotic stress tolerance (Zou et al. 2007, 2008). These findings suggested that the important roles of group A bZIPs in ABA and abiotic stress responses were conserved in flowering plants.
Despite the potential biotechnological utilization of group A AtbZIP orthologs in rice, relative few of them have been functionally studied in rice. Therefore, we carried out extensive expression analyses of this group of genes in four tissues of rice plants, including roots, panicles, seedlings and stems. We further analyzed the expression of the genes in response to several environmental stresses and plant hormones. In the context of this broad analysis, we chose OsbZIP72 from this group for detailed functional characterization. ABRE binding activity and transcription activation of OsbZIP72 were tested using yeast hybrid systems. Transgenic rice plants constitutively expressing OsbZIP72 showed hypersensitive to ABA and enhanced drought tolerance during seedling development. The results indicated that OsbZIP72 was an important regulator in abiotic stress response and ABA signaling transduction pathway. This was the first confirmed positive regulator of the drought tolerance among rice group A bZIPs.
Materials and methods
Identification and sequence analysis of the bZIP group A genes in rice
Sequences of Arabidopsis bZIP transcription factors previously identified (Deppmann et al. 2004) were downloaded from TAIR (http://www.arabidopsis.org/wublast/index2.jsp). Protein sequences of bZIP domains of eight Arabidopsis bZIPs representing the eight major groups of AtbZIPs (AtbZIP9, AtbZIP11, AtbZIP19, AtbZIP26, AtbZIP38, AtbZIP41, AtbZIP51, AtbZIP56) were used as queries to search against the protein database of rice with BLASTP in TIGR (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1). All hits with E-value below 0.001 were selected for further analysis. False hits were removed by manual inspection based on two criteria: (1) a well-defined basic region including a near invariant asparagine critical for sequence-specific DNA binding and (2) a canonical leucine zipper region with obvious amphipathic qualities (Deppmann et al. 2004). Multiple protein alignment was performed with ClustalX 1.81 using default parameters (Thompson et al. 1997). Phylogenetic tree was constructed with the neighbor-joining (NJ) method in ClustalX. Rice bZIPs locating within the same clade as group A AtbZIPs were identified as group A OsbZIPs.
Plant materials, growth condition and stress treatments
Oryza sativa cv. Nipponbare seeds (obtained from China National Rice Research Institute in Hangzhou, Zhejiang Province) were used in all experiment. Seeds germinated at 28°C in dark for 2 days were transferred to plant growth chamber to grow for 10 days under controlled conditions (12 h light 30°C/12 h dark 24°C cycles) to harvest the shoots and roots. Stems and panicles after heading were prepared from the plants grown in field. For stress and hormone treatments, 14-day-old seedlings were incubated in solutions containing 20% PEG-6000, 200 mM NaCl, 100 μM cis-, trans-ABA, 50 μM ACC (ethylene), 100 μM MeJA (methyljasmonate) and 10 μM NAA (naphthalene acetic acid) for 4 h and 12 h at 30°C, respectively. For cold treatment seedlings at the same developmental stage were treated at 4°C for 24 h and 48 h in dark. The pathogen used was Magnaporthe grisea, and the treatment was done according to previous study (Chen et al. 2006). These experiments were repeated twice, similar tendency was observed, expression levels shown in Fig. 2 were according to one time, error bars were based on three technical repeats. For the marker gene analysis, 14-day-old seedlings of WT control and of the transgenic rice lines were treated with/without 100 μM ABA for 5 h. RNAs were isolated from shoots.
Quantitative reverse transcription (RT)-PCR
Total RNAs of all the materials harvested were extracted using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The DNase-treated RNA was reverse-transcribed using M-MLV reverse transcriptase (Invitrogen). Quantitative PCR was performed on the Applied Biosystems 7500 real time PCR System using SYBR Premix Ex Taq™ (TaKaRa). The PCR thermal cycle conditions were as following: denature at 95°C for 10 s and 40 cycles for 95°C, 5 s; 60°C, 34 s. Two rice genes used as internal reference genes for calculating relative transcript levels were UBQ5 (AK061988) and eEF-1α (AK061464) (Jain et al. 2006). The primer efficiency used for calculating the relative quantification was 2.0 (Pfaffl 2001).
Yeast hybrid assay
For the DNA binding assay, the full open reading frame (ORF) of OsbZIP72 was generated by PCR (for primers, see Suppl. Table 1) and fused in frame with pGAD424 to construct pGAD-OsbZIP72. It was transformed into yeast strain YM4271 which was integrated with the reporter vector pHISi-4 × ABRE (5′ gtacgtgtc 3′) to determine the interaction of OsbZIP72 and ABRE. pHIS-4 × muABRE (5′ gtatatgtc 3′) with a 2 bp substitution at the center of the ABRE was used as a negative control. For transcription activation assay, sequencing confirmed PCR fragment of full ORF, C-terminal ORF with the bZIP domain and N-terminal ORF without bZIP domain were fused in frame with pGBKT7 to construct pGBKT7-OsbZIP72, pGBKT7-OsbZIP72ΔN, pGBKT7-OsbZIP72ΔC (Fig. 3b). pGBKT7 was used as a negative control, and the interaction between the BD-p53 and the AD-SV40 large T-antigen was used as a positive control (derived from Clontech). These different constructs were transformed into yeast strain AH109. The substrate used in the β-galactosidase assays was ONPG, and the analysis was carried out following the manufacturer’s specifications (Clontech).
Isolation of OsbZIP72, construction of overexpression vector, and gene transformation
The protein coding region of OsbZIP72 was amplified from seedling of O. sativa cv. Nipponbare by RT-PCR (for primers, see Suppl. Table 1). The sequencing-confirmed PCR fragment was ligated into the overexpression vector pCAMBIA1300S to construct pCAMBIA1300S-OsbZIP72. This construct was transformed into rice (Oryza sativa cv. Nipponbare) by Agrobacterium-mediated transformation method to generate transgenic rice plants (Hiei et al. 1994).
ABA sensitivity test of the transgenic plants
For the ABA sensitive test of the transgenic rice at the seedling stage, all the seeds were germinated on 1/2MS medium (Murashige and Skoog 1962) plates. Geminated rice plantlets of the same stage of transgenic rice and wild type (WT) control were transferred to 1/2MS medium with different concentrations of ABA (0, 2, 5 and 10 μM). Twelve days after grown in the green house with the temperature at 26°C, shoot height and root length were measured, and the root number was counted of each seedling.
Drought tolerance test
Transgenic seedlings were grown hydroponically for 15–18 days to 3-leaf stage, and parallel grown WT rice plants of the same stage were used as control. Whole plants were exposed to air for 9.5 h, then all the plants were rehydrated and recovered for an additional 10 days. The survival rates of each line and the WT control were calculated.
Germination test
For germination test, transgenic rice seeds and WT rice seeds were put on the 1/2MS medium with or without 10 μM ABA, the germinated seeds were counted every 12 h till most seeds were germinated.
Results
Identification of group A OsbZIPs
We found through the BLAST search a total of 85 loci in the rice genome as potential bZIPs, largely consistent with those identified in an earlier study (Nijhawan et al. 2008). Four genes, LOC_Os02g33560, LOC_Os11g05640, LOC_Os11g11100,LOC_Os12g09270, identified previously were exclude from our analysis because they failed to meet a more stringent standard that we used to define bZIP transcription factors (Deppmann et al. 2004). All OsbZIP domain sequences were aligned with those of AtbZIPs using ClustalX (Thompson et al. 1997). A neighbor-joining tree was constructed based on the alignment result (Suppl. Fig. S1). From the phylogenetic analysis, it was found that all the group A AtbZIPs were grouped into a distinct clade. The OsbZIPs contained within this clade were identified as rice orthologs of Arabidopsis group A bZIPs, and thereby were designated as group A OsbZIPs (Table 1). The names used in this study for all OsbZIPs were the same as in the previous report (Nijhawan et al. 2008).
Phylogenetic analysis of Arabidopsis and rice group A bZIPs
In order to better understand the relationships within the group A bZIPs of rice and Arabidopsis, the entire protein sequences of this group from rice and Arabidopsis were aligned and a bootstrap NJ-tree was constructed using ClustalX (Thompson et al. 1997) (Fig. 1). According to this phylogenetic tree, all of the bZIPs could be classified into three subgroups. Each subgroup contains both rice and Arabidopsis bZIPs, indicating that the divergence of these subgroups predated the divergence of dicots and monocots. It also indicated that the protein sequences of these bZIPs were well conserved between dicots and monocots.
Expression profiles of OsbZIP group A genes
Of the eleven rice genes, OsbZIP29 had no EST or full-length cDNA evidence, and could not be detected by qRT-PCR in our experiment. The expression analysis was done for the remaining ten genes. Expression patterns and levels of these OsbZIPs were rather different as illustrated by different scales of relative expression among the genes (Fig. 2). The expression levels of OsbZIP9, 10, 40, 42 and 62 were relatively low in all tissues, while expression levels of OsbZIP12, 23, 46, 66 and 72 were relatively high. As compared among different tissues of the same genes, the expression level of OsbZIP62 was relatively low in seedling; the expression levels of OsbZIP12 and 23 were relatively low in root; the expression levels of OsbZIP40 and 72 were relatively low in both root and stem; the expression levels of OsbZIP10 and 46 were relatively high in panicle.
Expression of ten rice group A OsbZIP genes were detected by qRT-PCR in response to stresses (PEG, NaCl, Cold and Blast) and hormones (ABA, ACC, MeJA and NAA) in rice seedlings. The time of the treatments were indicated. Seedling was used as control. The right column R root, S stem, P panicle. n = 3, error bars, ±SE
The expression pattern in response to environmental stresses and several hormones was also investigated. As shown in Fig. 2, the expression levels of all the bZIPs except OsbZIP42 were obviously unregulated in response to PEG, NaCl, Cold, ABA and ACC. The expression of OsbZIP10 was induced by pathogen treatment. The expression levels of OsbZIP9, 12, 23, 40, 46, 62, 66 and 72 were elevated by MeJA treatment. The expression levels of OsbZIP9, 10, 12, 23, 62 and 72 were elevated by NAA treatment. The expression levels of OsbZIP9, 23, 46 and 72 were repressed by pathogen treatment. The expression level of OsbZIP42 was repressed by PEG, NaCl, MeJA and NAA treatment. These differential expression patterns revealed specific and distinct roles of group A OsbZIPs in response to different environmental stresses.
A previous study investigated the expression of OsbZIPs under dehydration, salinity and cold stress treatments based on a microarray analysis (Nijhawan et al. 2008). The data revealed that 7 members of this group, OsbZIP10, 12, 23, 46, 62, 66 and 72, were induced by one or more of these stress treatments which was consistent with our results. However, three other OsbZIPs, OsbZIP9, OsbZIP40 and OsbZIP42, showed different expression profiles, they were induced in our study but not in the previous analysis (Nijhawan et al. 2008). This might be explained by the use of different rice cultivars (indica rice IR64 was previous tested) and/or somewhat different conditions of abiotic stress treatments between the two studies.
OsbZIP72 binds to ABRE and transactivates reporter genes in yeast
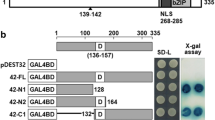
ABFs can activate the expression of downstream genes in response to ABA through binding to ABREs (Choi et al. 2000; Kang et al. 2002). In this study, we used the yeast one hybrid system to examine the binding activity of OsbZIP72 to ABRE. As shown in Fig. 3a, upper two of the four plates show yeast reporter strain integrated with pHIS-ABRE. This yeast reporter strain was transformed with pGAD-OsbZIP72 and the negative control vector pGAD424, respectively. They all grew well on the SD/His-Leu-plate. However, when 30 mM 3AT was added to the medium, yeast cells transformed with pGAD-OsbZIP72 still grew well, whereas the yeast cells transformed with pGAD424 control failed to grow. The lower two plates show yeast reporter strain integrated with pHIS-muABRE. This yeast reporter strain was also transformed with pGAD-OsbZIP72 and negative control vector pGAD424, respectively. They all grew well on the SD/His-Leu-plate. However, when 30 mM 3AT was added to the medium, they all failed to grow even for that transformed with pGAD-OsbZIP72. These results demonstrated that OsbZIP72 could specifically bind to ABRE in yeast.
ABRE binding activity analysis and transactivation tests in yeast. a ABRE binding activity analysis of OsbZIP72 by yeast one hybrid system. Sketch maps on the left show the construction of the vectors used in this experiment. Photographs on the right show the growth behavior of the transformants on the SD/His-Leu- plates with different concentrations of 3-AT. 1 pHIS-ABRE + pGAD424, 2 pHIS-ABRE + pGAD-OsbZIP72, 3 pHIS-muABRE + pGAD424, 4 pHIS-muABRE + pGAD-OsbZIP72. b The constructs of the plasmids pGBKT7-OsbZIP72, pGBKT7-OsbZIP72ΔN and pGBKT7-OsbZIP72ΔC were shown in the upper parts. Transactivation analysis of pGBKT7-OsbZIP72, pGBKT7-OsbZIP72ΔN and pGBKT7-OsbZIP72ΔC by yeast two hybrid analysis were detected on the SD/Trp- and SD/Trp-/His-/Ade-media, respectively. β-Gal activity presents the mean value of 4 independent measurements, and error bars indicate the standard variation. pGBKT7 was used as a negative control, and the interaction between the BD-p53 and the AD-SV40 large T-antigen was used as a positive control (derived from Clontech)
Yeast two hybrid system was used to determine the transactivation activity of OsbZIP72. All transformants grew well on the SD/Trp-plate (Fig. 3b). However, when cultured on the SD/Trp-/Adh-/His-plates, transformants of pGBKT7-OsbZIP72, pGBKT7-OsbZIP72ΔC still grew well whereas pGBKT7-OsbZIP72ΔN and the negative control failed to grow. These results were consistent with the β-galactosidase activity analysis, and indicated that OsbZIP72 had a transactivation activity, which was depending on its N-terminal region.
Expression analysis of the ABA signal pathway marker genes in the transgenic rice plants
Many ABA signal transduction components have been identified, including the ABF/ABRE system. The ABA-depended phosphorylation of the ABFs was indispensable to its activation. This phosphorylation process was performed by SnRK family protein kinases (Lopez-Molina et al. 2001; Furihata et al. 2006), and it could be inhibited by the PP2C-type protein phosphatases (Yoshida et al. 2006). The activated ABF/ABRE system could induce the expression of the late embryogenesis abundant (LEA) genes, which were proved to participate in the acquisition of desiccation tolerance (Mundy and Chua 1988; Xiao et al. 2007). As an ABRE-binding factor, OsbZIP72 may function downstream of SnRKs and PP2Cs and upstream of LEAs.
To test these predictions, we analyzed the expression levels of a PP2C family gene (Abi2, NM_001049950), a SnRK family gene (SAPK10, NM_001057188), and two LEA genes (LEA3, NM_001062730; Rab16, NM_001074376) in the transgenic plants under ABA treatment. Expression levels of all the marker genes analyzed were induced by ABA treatment both in WT control and our transgenic rice plants (Fig. 4). The expression levels of the upstream marker genes, Abi2 and SAPK10, did not show significant differences between control and the transgenic rice plants under the ABA treatment. But the expression levels of downstream marker genes, LEA3 and Rab16, were significantly higher in the transgenic rice plants than the control under ABA treatment. The results were consistent with the predictions and supported the role of OsbZIP72 as an ABRE-binding factor in the ABA signal transduction pathway.
Overexpression of OsbZIP72 makes seedlings hypersensitive to ABA
A total of 15 transgenic rice lines constitutively expressing OsbZIP72 under the control of the cauliflower mosaic virus 35S promoter were obtained. The integration of the construct into transgenic rice genome was confirmed by PCR using hygromycin specific primers (data not show), and the overexpression of OsbZIP72 in 13 transgenic rice lines was confirmed by qRT-PCR using OsbZIP72 specific primers (Suppl. Fig. S2). Using OsActin-1 as the reference gene, all of the transgenic lines showed higher expression levels of OsbZIP72 than the WT control.
We investigated the effect of ABA on the seedling development of the OsbZIP72 overexpression lines. Homozygous transgenic lines, 4 and 17 of the T2 generation were used to measure the ABA effect. There were no dramatic differences between WT and the transgenic rice plants compared in shoot length, root length and root number in medium without ABA. However, as shown in Fig. 5 the shoot length of WT growing on the medium with 2-μM ABA was reduced to 47.7% of the control growing on the medium without ABA, while shoot lengths of transgenic rice line 4–1 and line 17–1 were arrested to 15.0 and 12.0%, respectively. When the ABA concentrations in the medium were elevated to 5 and 10 μM, the shoot lengths of WT rice were reduced to 22.4 and 15.1% of control, respectively, while the shoot lengths of transgenic plants of both lines were arrested to about 5% of the control. The same tendencies were observed in root lengths and root numbers in response to ABA (Fig. 5). The severity of ABA induced growth arrest of the seedling length, root length and root number were more significant in the transgenic lines 4 and 17 than in the WT control (t test, P < 0.01). The root numbers of the WT rice plants were severely arrested when the ABA concentration was elevated to 10 μM, so they did not show significant difference with root numbers of the transgenic rice plants. The results indicated that overexpression of OsbZIP72 in transgenic rice plants caused the seedling development more sensitive to exogenous ABA, suggesting that OsbZIP72 was a positive regulator in the ABA signal transduction pathway.
Seedling development of OsbZIP72 overexpression transgenic rice plants under ABA treatment. a The WT control and transgenic rice lines L4 and L17 were grown under different concentrations of ABA (0, 2, 5, 10 μM) for 12 days, some representative plants were shown, the scale bar in this figure represented 5 cm. b Shoot and root lengths, root numbers at different ABA concentrations of the WT control (Nip) and the transgenic rice lines L4 and L17. Error bars represent the SE (n > 10). Bar diagram below illustrated the expression levels of OsbZIP72 of the WT control and the transgenic rice lines
Overexpression of OsbZIP72 enhances drought tolerance
Homozygous transgenic lines, 4, 12, and 17 of the T2 generation were used for drought tolerance test. Figure 6 showed that an average of 2.1% of the WT seedlings survived, while an average of 58.6% of line 4, 62.9% of line 12 and 57.1% of line 17 survived. The survival rates were significantly higher in the transgenic lines 4, line 12 and line 17 than in control (t test, P < 0.001).
Drought tolerance test of OsbZIP72-overexpressing rice plants. a Performance of WT control and OsbZIP72-overexpression line L4, L12 and L17 after 9.5 h drought stress and 10 days recovery. b Expression levels of OsbZIP72 in the WT control and the transgenic lines. c Survival rate of the WT and three transgenic rice lines. Error bars of the survival rates were based on three replicates (n > 30)
OsbZIP72 overexpression plants show germination hypersensitivity to ABA
Homozygous transgenic line 4 of the T2 generation was used for germination test. Figure 7a showed that seed germination of the transgenic rice was slower than that of the WT seeds on the plates with and without of ABA.
Discussion
The Arabidopsis bZIPs were divided into 10 groups. Seven members of the group A have been reported, and the functional information available suggested that most of them were involved in ABA and stress signaling (Jakoby et al. 2002). Homology search and phylogenetic analysis identified 11 genes that were potentially rice orthologs of the Arabidopsis group A bZIPs. If they did have similar functions with their Arabidopsis counterparts, these OsbZIPs could be valuable targets for biotechnological improvement of stress resistance of rice cultivars. Our expression analysis indeed confirmed that most of the rice genes were induced by environmental stresses and treatments of plant hormones including ABA and ethylene. These results thus suggested that group A OsbZIPs might be functionally associated with ABA signal pathway and stress responses.
Phylogenetic analysis showed that TRAB1 (OsbZIP66) formed a monophyletic group with three rice genes, OsbZIP23, OsbZIP46, and OsbZIP72 (Fig. 1), suggesting the possibility of functional conservation between them. The expression analysis showed that all of the four rice genes expressed at relatively high levels in all the tissues analyzed, and that their expression levels were induced by PEG, NaCl, Cold, ABA and ACC. Thus OsbZIP23, OsbZIP46, and OsbZIP72 could also be involved in the ABA signaling pathway as TRAB1. To further investigate the role of these genes in the ABA signaling pathway, we randomly chose OsbZIP72 for further experimentation.
Previous reports of TRAB1 and OsABI5, two OsbZIPs in group A, showed that they could bind to ABRE. Hobo et al. (1999) showed that TRAB1 could bind to Motif A (5′ TACGTGTC 3′), Em1a (5′ GACACGTGGC 3′) and Motif I (5′ TACGTGGC 3′) from the promoters of rice gene Osem (Hattori et al. 1995) and Rab16A (Skriver et al. 1991) and a wheat gene Em (Guiltinan et al. 1990), respectively. Yeast one hybrid assay showed that OsABI5 could bind to G-box element which also had the ABRE core sequence 5′ ACGT 3′ (Zou et al. 2007). We chose Motif A (5′ TACGTGTC 3′) as the ABRE binding target in this study. A mutant form muMotif A (5′ TATATGTC 3′), with 2 bp substitutions at the center of the motif, was used as a negative control. Yeast one hybrid experiment in this study showed that OsbZIP72 could specifically bind to ABRE element (5′ TACGTGTC 3′). This result indicated that OsbZIP72 could bind to the ABRE in promoters of the down stream genes and was involved in the ABA signaling pathway. In vivo analysis of TRAB1 showed that it could transactivate the downstream reporter genes in response to ABA application (Hobo et al. 1999). Yeast experiment of OsABI5 also showed that it had transcriptional activity, and its N-terminal region was necessary for its transcriptional activity. We found in our study that OsbZIP72 also had transcriptional activity, and its activity was also depending on its N-terminal.
Some of bZIPs in this group were proved to be involved in the ABA signaling pathway experimentally. Overexpression of Arabidopsis bZIP gene ABI5 and its rice orthologous gene OsABI5 in Arabidopsis made the transgenic plants more sensitive to exogenous ABA (Lopez-Molina et al. 2001; Zou et al. 2007). With transgenic rice plants overexpressing OsbZIP72, we tested the ABA sensitivity of the transgenic rice plants during seedling development. Our result showed that overexpression of OsbZIP72 also could make the transgenic rice plants hypersensitive to ABA, indicating that OsbZIP72 played a role in the ABA signaling transduction pathway during the seedling development.
We tested the expression levels of the ABA downstream genes of the transgenic rice plants. When treated with ABA, the transgenic lines had higher levels of transcripts of two LEA genes, which were known to be downstream of ABFs in the ABA signaling pathway, than control plants. However, the expression levels of these two genes did not show significant difference between transgenic rice and the WT when ABA was not present, indicating that OsbZIP72 alone was not sufficient to activate these two target genes and it probably required additional factor(s) for its gene activation function.
The expressions of Abi2 and SAPK10, which were known to be upstream of ABFs, did not show significant difference between transgenic and control plants. These evidences together strongly supported the function of OsbZIP72 as an ABF in the ABA signal transduction pathway.
Seed dormancy enables seed to avoid the unfavorable environmental condition and germinate when the conditions are convenient. ABA plays a crucial role in maintaining seed dormancy (Koornneef et al. 2002). Previous studies had shown that bZIP genes in this group might play important roles in the ABA induced seed germination arrestment (Lopez-Molina et al. 2001; Zou et al. 2007). In our work we observed that seed germination of transgenic plants overexpressing OsbZIP72 was delayed in transgenic rice line 4 than that of the WT rice on the plates with and without ABA (Fig. 7a). This was probably due to the elevated ABA sensitivity in the transgenic plants. The results suggested that OsbZIP72 might be also involved in the ABA signaling pathway in regulation of seed germination.
Drought is one of the most important environmental constraints of the rice productivity. The enhancement of drought tolerance by overexpression of several group A AtbZIPs has been reported (Kang et al. 2002; Oh et al. 2005). TRAB1 and OsABI5 are the only two rice orthologs of this group that have been functionally studied. There was no stress tolerance improvement report with regard to TRAB1, and OsABI5 was identified to be a negative regulator of drought and salt tolerance (Zou et al. 2008). OsbZIP72 is the first positive regulator identified in this group for rice drought tolerance. We observed that transgenic rice plants overexpressing OsbZIP72 were hypersensitive to ABA and had elevated expression of the ABA responsive genes, indicating it is a positive regulator in the ABA signal transduction pathway. This study thus has identified a regulatory gene that is potentially useful for engineering drought tolerant rice (Zhang et al. 2004; Umezawa et al. 2006).
Abbreviations
- bZIP :
-
Basic leucine zipper
- OsbZIP :
-
Oryza sativa bZIP
- AtbZIP :
-
Arabidopsis bZIP
- ABA:
-
Abscisic acid
- ABRE:
-
ABA responsive element
- ABF :
-
ABRE binding factor
- AREB:
-
ABRE binding protein
- ABI5 :
-
ABA insensitive 5
- RT-PCR:
-
Reverse-transcriptase polymerase chain reaction
- LEA :
-
Late embryogenesis abundant genes
- WT:
-
Wild type
References
Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425–435
Chen QH, Wang YC, Zheng XB (2006) Genetic analysis and molecular mapping of the avirulence gene PRE1, a gene for host-species specificity in the blast fungus Magnaporthe grisea. Genome 49:873–881
Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C (2004) Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res 32:3435–3445
Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103:1988–1993
Guiltinan MJ, Marcotte WR Jr, Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250:267–271
Hattori T, Terada T, Hamasuna S (1995) Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J 7:913–925
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96:15348–15353
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14:343–357
Kim SY, Chung HJ, Thomas TL (1997) Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J 11:1237–1251
Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5:33–36
Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98:4782–4787
Mundy J, Chua NH (1988) Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J 7:2279–2286
Mundy J, Yamaguchi-Shinozaki K, Chua NH (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA 87:1406–1410
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146:333–350
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10:296–302
Skriver K, Olsen FL, Rogers JC, Mundy J (1991) cis-Acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88:7266–7270
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 17:113–122
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115:35–46
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl):S165–S183
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281:5310–5318
Zhang JZ, Creelman RA, Zhu JK (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zou M, Guan Y, Ren H, Zhang F, Chen F (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360:307–313
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Acknowledgments
We thank Professors Zuhua He and Xiaoya Chen (Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for providing us the rice overexpression vector pCAMBIA1300S and the yeast experimental vectors, and Professor Tao Sang (Department of Plant Biology Michigan State University) for critical reading of the manuscript. This work was supported by the grants from the Ministry of Science and Technology of China (2006AA10A102), the Chinese Academy of Sciences (KSCW2-YW-N-024), and the National Natural Science Foundation of China (30821004).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lu, G., Gao, C., Zheng, X. et al. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615 (2009). https://doi.org/10.1007/s00425-008-0857-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0857-3