Abstract
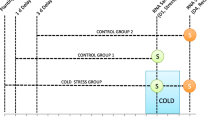
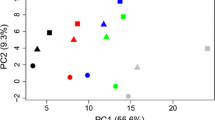
Our laboratory has been working toward increasing our understanding of the genetic control of cold hardiness in blueberry (Vaccinium section Cyanococcus) to ultimately use this information to develop more cold hardy cultivars for the industry. Here, we report using cDNA microarrays to monitor changes in gene expression at multiple times during cold acclimation under field and cold room conditions. Microarrays contained over 2,500 cDNA inserts, approximately half of which had been picked and single-pass sequenced from each of two cDNA libraries that were constructed from cold acclimated floral buds and non-acclimated floral buds of the fairly cold hardy cv. Bluecrop (Vaccinium corymbosum L.). Two biological samples were examined at each time point. Microarray data were analyzed statistically using t tests, ANOVA, clustering algorithms, and online analytical processing (OLAP). Interestingly, more transcripts were found to be upregulated under cold room conditions than under field conditions. Many of the genes induced only under cold room conditions could be divided into three major types: (1) genes associated with stress tolerance; (2) those that encode glycolytic and TCA cycle enzymes, and (3) those associated with protein synthesis machinery. A few of the genes induced only under field conditions appear to be related to light stress. Possible explanations for these differences are discussed in physiological context. Although many similarities exist in how plants respond during cold acclimation in the cold room and in the field environment, there are major differences suggesting caution should be taken in interpreting results based only on artificial, cold room conditions.





Similar content being viewed by others
Abbreviations
- CA:
-
Cold acclimated
- EST:
-
Expressed sequence tag
- NA:
-
Non-acclimated
References
Aase JK, Siddoway FH (1979) Crown-depth soil temperature and winter protection for winter wheat survival. Soil Sci Soc Am J 43:1229–1233
Ablett E, Seaton G, Scott K, Shelton D, Graham MW, Baverstock P, Slade Lee L, Henry R (2000) Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci 159:87–95
Adamska I (1997) ELIPs: light induced stress proteins. Physiol Plant 100:794–805
Alkharouf NW, Jamison DC, Matthews BF (2005) Online analytical processing (OLAP): a fast and effective data mining tool for gene expression databases. J Biomed Biotechnol 2:181–188
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipmann DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aro E-M, Virgin I, Andersson B (1993) Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta (BBA) Bioenerg 1143:113–134
Arora R, Rowland LJ, Panta GR (1997) Chill responsive dehydrins in blueberry: are they associated with cold hardiness or dormancy transitions? Physiol Plant 101:8–16
Arora R, Rowland LJ, Lehman JS, Lim CC, Panta GR, Vorsa N (2000) Genetic analysis of freezing tolerance in blueberry (Vaccinium section Cyanococcus). Theor Appl Genet 100:690–696
Arora R, Rowland LJ, Ogden EL, Dhanaraj AL, Marian CO, Ehlenfeldt MK, Vinyard B (2004) Dehardening kinetics, bud development, and dehydrin metabolism in blueberry cultivars during deacclimation at constant, warm temperatures. J Am Soc Hort Sci 129:667–674
Boatwright GO, Ferguson H, Sims JR (1976) Soil temperature around crown node influences early growth, nutrient-uptake, and nutrient translocation of spring wheat. Agron J 68:227–231
Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29:365–371
Browse J, Xin Z (2001) Temperature sensing and cold acclimation. Curr Opin Plant Biol 4:241–246
Byrne M, Murrell JC, Owen JV, Williams ER, Moran GF (1997) Mapping of quantitative trait loci influencing frost tolerance in Eucalyptus nitens. Theor Appl Genet 95:975–979
Chattopadhayay MK, Tiwari BS, Chattopadhayay G, Bose A, Sengupta DN, Ghosh B (2002) Protective role of exogenous polyamines on salinity-stressed rice (Oryza sativa) plants. Physiol Plant 116:192–199
Codd EF, Codd SB, Salley CT (1993) Providing OLAP (on-line analytical processing) to user-analysis: An IT mandate. Technical Report, EF Codd & Associates
Conley TR, Peng H-P, Shih M-C (1999) Mutations affecting induction of glycolytic and fermentative genes during germination and environmental stresses in Arabidopsis. Plant Physiol 119:599–608
Coulson SJ, Hodkinson ID, Strathdee AT, Block W, Webb NR, Bale JS, Worland MR (1995) Thermal environments of arctic soil organisms during winter. Arct Alp Res 27:364–370
Dhanaraj AL, Slovin JP, Rowland LJ (2004) Analysis of gene expression associated with cold acclimation in blueberry floral buds using expressed sequence tags. Plant Sci 166:863–872
Dhanaraj AL, Slovin JP, Rowland LJ (2005) Isolation of a cDNA clone and characterization of expression of the highly abundant, cold acclimation-associated 14 kDa dehydrin of blueberry. Plant Sci 168:949–957
Dolferus R, Jacobs M, Peacock W, Dennis E (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 105:1075–1087
Ezhova TA, Soldatova OP, Kalinina AI, Medvedev SS (2000) Interaction of ABRUPTUS/PINOID and LEAFY genes during floral morphogenesis in Arabidopsis thaliana (L.) Heynh. Genetika 36:1682–1687
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Gonorazky AG, Regente MC, de la Canal L (2005) Stress induction and antimicrobial properties of a lipid transfer protein in germinating sunflower seeds. J Plant Physiol 162:618–624
Guy CL (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Ann Rev Plant Physiol Plant Mol Biol 41:187–223
Guy C (1999) Molecular responses of plants to cold shock and cold acclimation. J Mol Microbiol Biotechnol 1:231–242
Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genetics 1:179–196
Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J (2000) A concise guide to cDNA microarray analysis. Biotechniques 29:548–556
Howe GT, Saruul P, Davis J, Chen THH (2000) Quantitative genetics of bud phenology, frost damage, and winter survival in an F2 family of hybrid poplars. Theor Appl Genet 101:632–642
Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M (2003) Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA 100:4921–4926
Jermstad KD, Bassoni DL, Wheeler NC, Anekonda TS, Aitken SN, Adams WT, Neale DB (2001) Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. II. Spring and fall cold-hardiness. Theor Appl Genet 102:1152–1158
Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136:4159–4168
Khan R, Alkharouf N, Beard HS, MacDonald M, Chouikha I, Meyer S, Grefenstette J, Knap H, Matthews BF (2004) Resistance mechanisms in soybean: gene expression profile at an early stage of soybean cyst nematode invasion. J Nematol 36:241–248
Kreps JA, Wu Y, Chang H-S, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Lebedeva OV, Ondar UN, Penin AA, Ezhova TA (2005) Effect of the ABRUPTUS/PINOID gene on expression of the LEAFY gene in Arabidopsis thaliana. Genetika 41:559–565
Leep RH, Andresen JA, Jeranyama P (2001) Fall dormancy and snow depth effects on winterkill of alfalfa. Agron J 93:1142–1148
Levi A, Panta GR, Parmentier CM, Muthalif MM, Arora R, Shanker S, Rowland LJ (1999) Complementary DNA cloning, sequencing, and expression of an unusual dehydrin from blueberry floral buds. Physiol Plant 107:98–109
Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcription factor using two microarray systems. Plant J 38:982–993
Mellander PE, Bishop K, Lundmark T (2004) The influence of soil temperature on transpiration: a plot scale manipulation in a young Scots pine stand. Forest Ecol Manage 195:15–28
Meyer G, Kloppstech K (1984) A rapidly light-induced chloroplast protein with a high turnover coded for by pea nuclear DNA. Eur J Biochem 138:201–207
Minhas D, Grover A (1999) Transcript levels of genes encoding various glycolytic and fermentation enzymes change in response to abiotic stress. Plant Sci 146:41–51
Moore JN (1993) The blueberry industry of North America. Acta Hort 346:15–26
Muthalif MM, Rowland LJ (1994) Identification of dehydrin-like proteins responsive to chilling in floral buds of blueberry (Vaccinium, section Cyanococcus). Plant Physiol 104:1439–1447
Rintamaki E, Kettunen R, Aro E-M (1996) Differential D1 dephosphorylation in functional and photodamaged photosystem II centers: dephosphorylation is a prerequisite for degradation of damaged D1. J Biol Chem 271:14870–14875
Rowland LJ, Panta GR, Mehra S, Parmentier-Line C (2004) Molecular genetic and physiological analysis of the cold-responsive dehydrins of blueberry. J Crop Improv 10:53–76
Rowland LJ, Ogden EL, Ehlenfeldt MK, Vinyard B (2005) Cold hardiness, deacclimation kinetics, and bud development among 12 diverse blueberry genotypes under field conditions. J Am Soc Hort Sci 130:508–514
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13:61–72
Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotech 7:161–167
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Biotech 3:217–223
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Ann Rev Plant Physiol Plant Mol Biol 50:573–599
Thomashow M (2001) So what’s new in the field of plant cold acclimation? Lots! Plant Physiol 125:89–93
Wei H, Dhanaraj AL, Rowland LJ, Fu Y, Krebs SL, Arora R (2005) Comparative analysis of expressed sequence tags from cold-acclimated and non-acclimated leaves of Rhododendron catawbiense Michx. Planta 221:406–416
Weiser CJ (1970) Cold resistance and injury in woody plants. Science 169:1269–1278
Wi SJ, Park KY (2002) Antisense expression of carnation cDNA encoding ACC synthase or ACC oxidase enhances polyamine content and abiotic stress tolerance in transgenic tobacco plants. Mol Cells 13:209–220
Wi SJ, Kim WT, Park KY (2006) Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep. DOI 10.1007/s00299-006-0160-3
Wilkins TA, Smart LB (1996) Isolation of RNA from plant tissue. In: Krieg PA (ed) A laboratory guide to RNA: isolation, analysis, and synthesis. Wiley, New York, pp 21–41
Xin Z, Browse J (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23:893–902
Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238:17–25
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization of cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acid Res 30(4):e15
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Dhanaraj, A.L., Alkharouf, N.W., Beard, H.S. et al. Major differences observed in transcript profiles of blueberry during cold acclimation under field and cold room conditions. Planta 225, 735–751 (2007). https://doi.org/10.1007/s00425-006-0382-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0382-1