Abstract
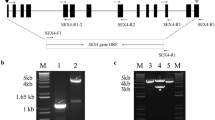
A full-length coding domain sequence of a gene analogous to granule-bound starch synthase (GBSS; ADP-glucose-starch glucosyltransferase, EC 2.4.1.21) was cloned and defined as OsGBSSII based on a Nitrogen (N)-starvation-induced cDNA library constructed using the rapid subtraction hybridization method. The deduced amino acid sequence of OsGBSSII was 62–85% identical to those of GBSS proteins from other plant species. The exon/intron organization of OsGBSSII was similar to that of OsGBSSI. OsGBSSII was mainly expressed in leaves and its protein was exclusively bound to starch granules in rice leaves, which suggests that the amylose in rice leaves is synthesized by OsGBSSII. N-starvation-induced expression of OsGBSSII could be repressed by supplying nitrate, ammonia or amino acid (glutamic acid or glutamine), glucosamine (an inhibitor of hexokinase) or dark conditions. These results indicate that N-starvation induction was dependent on the photosynthetic product and hexokinase in rice leaves. Sugars induced the accumulation of OsGBSSII transcripts in excised leaves through glycolysis-dependent pathways. OsGBSSII gene expression is regulated by the circadian rhythm in rice leaves.









Similar content being viewed by others
Abbreviations
- AGPase:
-
ADP-glucose pyrophosphorylase
- BE:
-
branching enzyme
- C:
-
Carbon
- GBSS:
-
granule-bound starch synthase
- Glc:
-
glucose
- Gln:
-
glutamine
- Glu:
-
glutamic acid
- HXK:
-
hexokinase
- N:
-
Nitrogen
- RaSH:
-
Rapid Subtraction Hybridization
- Suc:
-
sucrose
References
Blakeney AB, Matheson NK (1984) Some properties of the stem and pollen starches of rice. Starch 36:265–269
Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120:205–216
Denyer K, Barber LM, Edwards EA, Smith AM, Wang TL (1997) Two isoforms of the GBSSI class of granule-bound starch synthase are differentially expressed in the pea plant (Pisum sativum L.). Plant Cell Environ 20:1566–1572
Edwards A, Vincken JP, Suurs LCJM, Visser RGF, Zeeman S, Smith A, Martin C (2002) Discrete forms of amylose are synthesized by isoforms of GBSSI in pea. Plant Cell 14:1767–1785
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Furukawa K, Tagaya M, Tanizawa K, Fukui T (1993) Role of the conserved Lys-X-Gly-Gly sequence at the ADP-glucose-binding site in Escherichia coli glycogen synthase. J Biol Chem 268:23837–23842
Geiger DR, Servaites JC (1994) Diurnal regulation of photosynthetic carbon metabolism in C3 plants. Annu Rev Plant Physiol Plant Mol Biol 45:235–256
Harn C, Knight M, Ramakrishnan A, Guan H, Keeling PL, Wasserman BP (1998) Isolation and characterization of the zSSIIa and zSSIIb starch synthase cDNA clones from maize endosperm. Plant Mol Biol 37:639–649
Hirano HY, Tabayashi N, Matsumura T, Tanida M, Komeda Y, Sano Y (1995) Tissue-dependent expression of the rice wx gene promoter in transgenic rice and petunia. Plant Cell Physiol 36:37–44
Jiang H, Kang DC, Alexandre D, Fisher PB (2000) RaSH, a rapid subtraction hybridization approach for identifying and cloning differentially expressed genes. Proc Natl Acad Sci USA 97:12684–12689
Jiang HW, Dian WM, Liu FY, Wu P (2003a) Isolation and characterization of two fructokinase cDNA clones from rice. Phytochemistry 62:47–52
Jiang HW, Dian WM, Wu P (2003b) Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 63:53–59
Konishi Y, Nojima H, Okuno K, Asaoka M, Fuwa H (1985) Characterization of starch granules from waxy, nonwaxy, and hybrid seeds of Amaranthus hypochondriacus L. Agric Biol Chem 49:1965–1971
Kossmann J, Abel GJW, Springer F, Lloyd JR, Willmitzer L (1999) Cloning and functional analysis of a cDNA encoding a starch synthase from potato (Solanum tuberosum L.) that is predominantly expressed in leaf tissue. Planta 208:503–511
Li B, Geiger DR, Shieh WJ (1992) Evidence for circadian regulation of starch and sucrose synthesis in sugar beet leaves. Plant Physiol 99:1393–1399
Mérida A, Rodríguez-Galán JM, Vincent C, Romero JM (1999) Expression of the granule-bound starch synthase I (Waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant Physiol 120:401–409
Nakamura T, Vrinten P, Hayakawa K, Ikeda J (1998) Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol 118:451–459
Nakata PA, Okita TW (1995) Differential regulation of ADP-glucose pyrophosphorylase in the sink and source tissues of potato. Plant Physiol 108:361–368
Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol 38:655–662
Salehuzzaman SNIM, Jacobsen E, Visser RGF (1994) Expression patterns of two starch biosynthetic genes in vitro cultured cassava plants and their induction by sugars. Plant Sci 98:53–62
Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9:783–798
Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2:410–418
Smith AM, Denyer, K, Martin C (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48:67–87
Sokolov LN, Dejardin A, Kleczkowski LA (1998) Sugars and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase genes in Arabidopsis thaliana (thale cress). Biochem J 336:681–687
Umemura T, Perata P, Futsuhara Y, Yamaguchi J (1998) Sugar sensing and α-amylase gene repression in rice embryos. Planta 204:420–428
Visser RG, Stolte A, Jacobsen E (1991) Expression of a chimaeric granule-bound starch synthase-GUS gene in transgenic potato plants. Plant Mol Biol 4:691–699
Vrinten PL, Nakamura T (2000) Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol 122:255–263
Wang SJ, Yeh KW, Tsai CY (2001) Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci 161:635–644
Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J, Fujii F, Shomura A, Ando T, Kono I, Waki K, Yamamoto K, Yano M, Matsumoto T, Sasaki T (2002) A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14:525–535
Yamanouchi H, Nakamura Y (1992) Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol 33:985–991
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. International Rice Research Institute, Manila Philippines, pp 61–64
Zabawinski C, Van Den Koornhuyse N, D'Hulst C, Schlichting R, Giersch C, Delrue B, Lacroix JM, Preiss J, Ball S (2001) Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J Bacteriol 183:1069–1077
Acknowledgements
This research was supported by the National Nature Science Foundation of China (39830250), and the Natural Science Foundation of Zhejiang Province, China (ZA0106).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dian, W., Jiang, H., Chen, Q. et al. Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm. Planta 218, 261–268 (2003). https://doi.org/10.1007/s00425-003-1101-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-003-1101-9