Abstract
The microenvironment of proliferative and aggressive tumours, such as the brain tumour glioblastoma multiforme (GBM), is often acidic, hypoxic, and nutrient deficient. Acid-sensing ion channels (ASICs) are proton-sensitive Na+ channels that have been proposed to play a role in pH sensing and in modulation of cancer cell migration. We previously reported that primary glioblastoma stem cells (GSCs), which grow as multicellular tumour spheroids, express functional ASIC1a and ASIC3, whereas ASIC2a is downregulated in GSCs. Using a 2.5D migration assay, here we report that acidic pH dramatically increased migration of GSCs of the pro-neural subtype. Pharmacological blockade as well as CRISPR-Cas9-mediated gene knock-out of ASIC1a or stable overexpression of ASIC2a, however, revealed that neither ASIC1a nor ASIC3, nor downregulation of ASIC2a, mediated the aggressive migration at acidic pH. Therefore, we tested the role of two other proteins previously implicated in cancer cell migration: the Ca2+-activated K+ channel KCa3.1 (KCNN4) and phosphoinositide 3-kinase (PI3K). While pharmacological blockade of KCa3.1 did also not affect migration, blockade of PI3K decreased migration at acidic pH to control levels. In summary, our study reveals a strongly enhanced migration of GSCs at acidic pH in vitro and identifies PI3K as an important mediator of this effect.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma, IDH wild-type multiforme (GBM) is a type of glioma. GBM is the most common primary malignant brain tumour, with a survival rate of < 10% 5 years after the initial diagnosis [53]. Surgical resection, followed by radiotherapy and chemotherapy, is the most common treatment [46]; however, due to the infiltration of GBM into the surrounding brain tissue, the likelihood of tumour recurrence and eventual radio- and chemoresistance is > 86% [37]. GBM is a rapidly proliferating, aggressive, invasive, and undifferentiated type of tumour [17, 53] which uses glycolysis for ATP production, despite oxygen being available for oxidative phosphorylation [38]. This aerobic glycolysis, termed the Warburg effect, together with insufficient blood supply, acidifies the tumour microenvironment via the fermentation of pyruvate to lactate. An acidic microenvironment has been recognized to be relevant for proliferation, survival, metabolic adaptation, and migration in cancer [45, 51]. However, the mechanism of enhanced migration of cancer cells at acidic pH is insufficiently understood [7].
Acid-sensing ion channels (ASICs) are proton-gated Na+ channels, which, in the CNS, assemble either as homomeric ASIC1a or heteromeric ASIC1a/2a or ASIC1a/2b [52]. It has been shown that functional homomeric ASIC1a is also expressed in glioblastoma stem cell lines (GSCs) [47]. These GSCs grow as tumour spheres and are cultured without serum, because serum causes irreversible differentiation of GSCs [12]. GSCs closely recapitulate the genotype, gene expression pattern, and in vivo biology of primary tumours [29], whereas serum-cultured glioma cell lines do not [29, 30]. ASIC2a, in contrast, is downregulated in GSCs and in GBM tissue [47]. Thus, it is conceivable that ASIC1a senses acidosis in GBM. ASICs have indeed been implicated in pH sensing and migration of glioma cells [24, 40, 43, 49]. However, previous assays assessing the role of ASICs in migration in GBM cell cultures have been performed with serum-cultured glioma cell lines using transwell or wound-healing assays [24, 40, 43, 49].
Phosphoinositide 3-kinases (PI3K) are members of the PI3K/Akt/MTOR pathway, which is often mutated in cancer cells. PI3K is also known to be activated by hypoxia [55] and is linked to proliferation, cell survival, differentiation, migration, and poor disease prognosis in patients [32]. KCa3.1 (KCNN4) is an intermediate-conductance Ca2+-activated K+ channel (also known as IKCa) activated by intracellular Ca2+. It contributes to controlling the cell membrane potential [25] and has been reported to play a role in migration and proliferation of GBM [6, 42], hepatocellular carcinoma [31, 33], and migration in colorectal cancer [27], lung adenocarcinoma [54], and pancreatic cancer cells [18].
Here, we established a 2.5D migration assay and show much more aggressive migration of GSCs at an acidic extracellular pH (pHe). While we found no evidence for the involvement of ASICs or KCa3.1 in enhanced migration, blockade of PI3K abrogated enhanced migration at acidic pHe, identifying PI3K as a novel mediator of the more aggressive migration at acidic pHe.
Results and discussion
Characterization of pH-dependent migration in multicellular tumour GSCs
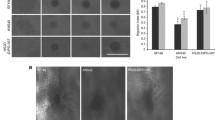
We established a sphere migration assay, which allowed us to test the migration of R54 cells, CD133+, pro-neural-like GSCs [35], in medium with a pH of 7.4 or 6.6. An acidic pH of 6.6 was chosen, as it is a relatively low pH value that can still be reached in GBM tissue [13]; in addition, it leads to robust activation of ASIC1a [2, 50]. Migration of GSCs out of spheres strongly increased at acidic pHe (Fig. 1a; p < 0.001). Culturing the spheres under normoxic or hypoxic (< 3% O2) conditions did not affect migration (Fig. 1b), which was unexpected, as numerous publications reported increased migration under hypoxic conditions [9, 20, 23]. It is possible that this was because the cores of the spheres were already hypoxic. Moreover, sphere sizes were the same between conditions (Fig. 1c). Thus, the profound increase in migration at pH 6.6 was driven by extracellular acidity and not by hypoxia, allowing for the specific assessment of acidity on migration.
Sphere migration strongly increases at pH 6.6, independent of oxygenation. a Representative pictures of 7-day-old spheres incubated at pH 7.4 or pH 6.6 on laminin-coated plates, with pictures taken every three hours. Scale bars = 200 µm. b Comparison of sphere migration under hypoxia (O2 < 3%) and normoxia. n (from left to right) = 176, 96, 221, 165 individual spheres from 12 biological replicates for hypoxia and three biological replicates for normoxia. ***p < 0.001 (Tukey’s test). c Sphere diameters. n (from left to right) = 109, 30, 93, 27. For sphere diameters, we did not account for variations in the number of technical replicates. Bars represent mean ± SD
ASIC1, ASIC2, and ASIC3 do not modulate pH-dependent migration
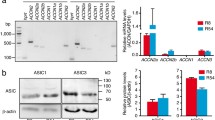
We assessed the expression of ASIC1, ASIC2, ASIC3, and ASIC4 genes using quantitative real-time PCR (qPCR). We found that ASIC1 was most strongly expressed and that ASIC3 and ASIC4 were expressed at two- to tenfold lower levels (Fig. 2), whereas ASIC2 was not expressed. These findings are in accordance with those of a previous study [47] and microarray data from a large cohort of glioma samples [14]. We did not find significant differences in expression in R54 cells maintained at pH 6.6 for 3 days (Fig. 2). We also tested mRNA expression in a recently described R54 cell line with an ASIC1a knockout [5] (Fig. 2). Compared to WT R54 cells, ASIC1 signal was still present, but was significantly lower (p = 0.0086) in ASIC1a knockout cells, likely due to the knock-out inducing a frameshift in the second exon of ASIC1a, thereby impairing the binding of the qRT-PCR probe at the exon junction of exon 2 to exon 3.
RNA expression of ASICs, stemness, and differentiation markers in R54 cells. Expression of ASIC1, ASIC3, ASIC4, stemness marker nestin, and the differentiation markers GFAP, TUBB3, and GALC, normalized to the expression of GAPDH. Dots represent biological replicates, which were at least n = 3 for every condition, with three technical replicates for each biological replicate. Bars represent mean ± SEM. GOI, gene of interest. **p < 0.01 (two-sided t test of ΔCt-values)
We also assessed the expression of the stemness marker nestin and the differentiation markers glial fibrillary acidic protein (GFAP), tubulin beta 3 (TUBB3), and galactocerebrosidase (GALC), which are markers of astrocytes, neurons, and oligodendrocytes, respectively. While nestin expression was high, the expression of TUBB3 and, in particular, of GFAP and GALC was much lower. There was a tendency for increased expression of nestin and of GFAP at pH 6.6, but this change was not statistically significant (p = 0.65 and p = 0.14, respectively) (Fig. 2).
To test for the involvement of ASIC1a and ASIC3 in enhanced migration at acidic pHe, we assessed migration of GSCs at acidic pH 6.6 in the presence of the potent ASIC1a inhibitor psalmotoxin 1 (PcTx1; 100 nM) or the potent ASIC3 inhibitor APETx2 (500 nM). Neither inhibitor had an effect on migration, irrespective of pH (Fig. 3a). They also did not affect sphere diameters (Fig. 3b). We then used the potent ASIC1 agonist MitTx (20 nM) to test whether the pharmacological activation of ASIC1 could enhance migration at neutral pHe. Like PcTx1, however, MitTx had no effect on migration (Fig. 3c) or on sphere diameters (Fig. 3d). Finally, we assessed the outgrowth of spheres using two independent R54 lines with an ASIC1a knockout [5]. Also ASIC1a knockout did not affect migration (Fig. 3e) or sphere diameters (Fig. 3f). These results consistently show that ASIC1a does not affect migration of R54 GSCs, neither at pH 7.4 nor at pH 6.6.
Enhanced migration of GSCs at acidic pH is not mediated by ASIC1a or ASIC3. a Inhibition of ASIC1 with 100 nM PcTx1 or of ASIC3 with 500 nM APETx2 did not affect migration at pH 7.4 or pH 6.6. n (from left to right) = 38, 28, 37, 29, 39, 33 individual spheres. b Sphere diameters between conditions were not significantly different from each other. c Activation of ASIC1a with 20 nM MitTx did not increase migration at pH 7.4 or pH 6.6. n (from left to right) = 14, 9, 13, 15. d Sphere diameters between conditions were not significantly different from each other. e Monoclonal R54 ASIC1a/b knockout cell lines migrated more aggressively at pH 6.6 than at pH 7.4. n (from left to right) = 26, 28, 22, 21, 29, 33. f Sphere diameters were not significantly different from each other. All experiments were performed in three biological replicates. Dots represent technical replicates. Bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 (Tukey’s test)
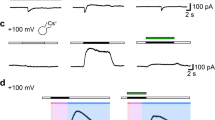
The downregulation of ASIC2a in GBM is striking [47], and it has been proposed that ASIC2a modulates migration of tumour cells [49, 57]. Therefore, we generated stably expressing ASIC2a R54 cells via lentiviral transduction. Electrophysiological characterization of the ASIC2a-overexpressing cells confirmed that ASIC currents in almost all cells had decreased proton sensitivity and an insensitivity to PcTx1 typical for heteromeric ASIC1a/ASIC2a [22] (Fig. 4a–e), confirming plasma membrane expression of ASIC2a in these cells. Strikingly, however, ASIC2a-overexpressing R54 cells displayed the same outgrowth as the wild-type parental cells and still aggressively migrated at an acidic pH 6.6 (Fig. 4f); sphere diameters were also unaffected (Fig. 4g). Thus, ASIC2a overexpression had no effect on migration of R54 GSCs, suggesting that ASIC2a downregulation by GBM cells did not affect migration.
Overexpression of ASIC2a does not change aggressive migration of GSCs at acidic pH. a Pie chart indicating the relative occurrence of R54 cells with no ASIC current, ASIC currents elicited by pH 6 and by pH 5, and ASIC currents elicited only by pH 5. n = 9 for WT and n = 10 for ASIC2-over-expressing (oe) cells. b Left, representative current traces of wild-type and ASIC2 oe cells at pH 6 and pH 5. Right, ratio of ASIC currents elicited by pH 5 and pH 6. Error bars represent the mean ± SD; the mean value for ASIC2 oe cells was 9.23. n = 7–9. c Left, representative current traces of wild-type and ASIC2 oe cells at pH 6, in the absence and presence of PcTx1. Right, ratio of ASIC currents elicited by pH 6, in the presence of PcTx1. Error bars represent the mean ± SD; n = 7–8. d Sphere migration of R54 wild-type and ASIC2 oe cells. n (from left to right) = 99, 184, 157, 83. The experiment was performed in three biological replicates. Dots represent technical replicates. Bars represent mean ± SD. ***p < 0.001 (Tukey’s test). e Sphere diameters between conditions were not significantly different from each other. n = 30 for each condition
PI3K, but not K Ca 3.1, modulates pH-dependent migration
So far, we only tested the influence on migration of acutely changing pHe from 7.4 to 6.6. To test whether chronic exposure to acidic pHe will also affect migration of GSCs, we maintained spheres for 2 weeks either at pH 7.4 or at pH 6.6 before performing the outgrowth assay for 6 h at either pH 7.4 or pH 6.6 (Fig. 5a). The large and abrupt change in pHe might favour selection of GSCs surviving at low pH rather than adaptation of GSCs to low pH [3]. Migration of cells from spheres that were maintained in acidic medium for 2 weeks was indeed strongly enhanced even when tested at pHe of 7.4 (p < 0.001; Fig. 5b). In fact, migration of these cells at pH 7.4 was not different from that of cells maintained at pH 7.4 and tested at pH 6.6. However, cells maintained at pH 6.6 and tested at an acute pHe of 6.6 showed the most aggressive migration (p < 0.001; Fig. 5b). Incubation of GSCs at an acidic pH might have primed them for aggressive migration or might have selected GSCs that migrated more aggressively. Sizes of spheres grown at pH 6.6 were smaller than those of spheres grown at pH 7.4 (Fig. 5b–c), which was expected, as the doubling time in pH 6.6 is longer for R54 GSCs [5]. It is unlikely, however, that the smaller sphere sizes affected the interpretation of this experiment, because, if anything, fewer cells should grow out of smaller spheres.
Acid-selection primes GSCs for aggressive migration and the PI3K-inhibitor wortmannin inhibits acute induction of migration by acidic pHe. a Experimental design of preincubation and assay pH conditions. Cells were grown into spheres for 14 days at either pH 7.4 or pH 6.6, and then positioned on laminin-coated wells for 6 h at pH 7.4 or pH 6.6. b Sphere migration with and without TRAM-34 and wortmannin. Experiments were performed in three biological replicates. Dots indicate technical replicates. n (from left to right) = 110, 141, 213, 208, 194, 191, 232, 101, 162, 119, 147, 198. Bars represent mean ± SD. ***p < 0.001 (Tukey’s test). c Sphere diameters. Dots indicate technical replicates. n (from left to right) = 131, 139, 159, 165, 132, 138, 136, 133, 125, 131, 162, 121. **p < 0.01, ***p < 0.001 (one-way ANOVA)
We then considered other targets affecting the pH-dependent migration of GSCs, and turned to KCa3.1, which has been reported to play a role in glioma cell migration [4, 6, 16, 42] and GBM invasiveness in vivo [6], and to PI3K, which is a well-known modulator of migration in GBM [1, 20, 21, 34]. KCa3.1 has been reported to be overexpressed in patient-derived GBM [48], whereas PI3K is overexpressed in GBM cell lines but not in patient-derived samples [56]. To assess the role of KCa3.1 and PI3K, we added the KCa3.1 inhibitor TRAM-34 (10 µM) or the pan-PI3K inhibitor wortmannin (1 µM) to the spheres 30 min before the start of the migration assay; we performed these assays with spheres maintained at pH 7.4 or at pH 6.6. While TRAM-34 did not affect migration under any conditions, wortmannin strongly reduced migration, but only when outgrowth was tested at pHe 6.6 (Fig. 5b). In contrast, enhanced outgrowth at pHe 7.4 of cells maintained at pH 6.6 was not reduced by wortmannin (p = 0.51). This suggests that PI3K plays an essential role in the acute induction of migration by acidic pHe and that the priming of migration or selection of aggressively migrating GSCs by acidic pH is PI3K-independent and relies on a different mechanism, at least when tested at pH 7.4. The finding that wortmannin strongly reduced migration at pHe 6.6 of cell maintained at acidic pH below the level of migration of these cells at pHe 7.4 is not entirely consistent with this interpretation and suggests a complex interplay of different mechanisms in acid-dependent migration of GSCs.
We note that the 2.5D sphere migration assay can only partially reproduce migration of tumour cells in situ. Cancer cells in situ are likely to be confronted with slower changes in pHe than the abrupt changes used in our experiments. In addition, serum-free conditions may not accurately represent the micro-environment of tumours in vivo. Nevertheless, we believe that our assay provides an important addition to earlier migration assays assessing the role of ASICs in tumour cell migration, which used serum-cultured glioma cell lines, which do not well represent the parental tumour [29, 30], in combination with wound-healing or transwell migration assays. The exact effect of acidic condition on glioblastoma cancer stem cell migration in vivo warrants further investigation.
Our findings using patient-derived R54 GSCs and a 2.5D migration assay do not confirm previous publications reporting that ASIC1, ASIC2, or ASIC3 play a role in pH- or hypoxia-dependent migration in CNS [24, 40, 43, 49], non-CNS tumours [57], and normal tissues [15]. In this respect, it is noteworthy that some of these studies assessed migration only at pH 7.4 [15, 24, 40, 49]. Mechanistically, it is unclear how a proton-gated ion channel can influence migration at neutral pH. Indeed, a recent study could not reproduce inhibition of migration by PcTx1 at pH 7.4 in U87MG GBM cells [43], as it had previously been reported for the unspecific ASIC blocker amiloride [49]. However, the same study found enhanced migration at pH 7.0, which could be inhibited by PcTx1 in two GBM cell lines [43]. Similarly, another study found an inhibition of enhanced migration at pH 6.4 by amiloride or by siRNA targeting ASIC1a or ASIC3 in two pancreatic cancer cell lines [58]. Moreover, overexpression of ASIC2a increased migration of colorectal cancer cells at pH 6.5, whereas ASIC2 knockdown decreased migration. Thus, there is conflicting evidence regarding the role of specific ASICs in cancer cell migration. This conflicting evidence could be due to differences in pH used in these studies [45] or to a different role of ASICs in cell lines originating from different tumours. Our study shows that ASICs do not have a universal role in migration of tumour cells.
Likewise, a role for KCa3.1 has been reported in numerous studies [6, 16, 27, 31, 42, 48] but could not be reproduced by us in a 2.5D sphere migration assay in R54 GSCs. This might be due to the fact that R54 GSCs are of the pro-neural subtype [35], while it has been proposed that KCa3.1 is a marker for the mesenchymal subtype of GSCs [26]. It will be worthwhile to assess in the future whether KCa3.1 plays a role in acid-induced migration of mesenchymal subtype GSCs.
In contrast, our findings reveal a novel and prominent role for PI3K in acid-induced migration of R54 GSCs. Because PI3K is an intracellular enzyme, it must have been activated either by a non-ASIC proton-sensor on the surface of R54 cells or by a drop in intracellular pH that often accompanies extracellular acidification. A few previous studies already reported an activation of PI3K by acidic pHe: activation of PI3K is crucial for triggering fatty acid synthesis in liver cancer cells by an acidic TME [36], for activation of human neutrophils by extracellular acidosis [8], and for cardioprotection by acidic reperfusion [39]. The precise mechanisms of PI3K activation and its downstream targets in R54 cells need to be further explored in the future.
Conclusion
In this study, using a 2.5D sphere migration assay and patient-derived R54 GSCs, we could not confirm a role for ASIC1, ASIC2, ASIC3, or KCa3.1 in pH-dependent migration of GBM cells. However, wortmannin reduced migration induced by acute extracellular acidosis suggesting that PI3K plays a crucial role in pH-dependent migration of GBM cells. The mechanism by which acidic pHe activates PI3K to induce migration remains unclear and needs to be elucidated in the future.
Methods
Cell culture
The cell line R54 [35] is an IDH1/2 WT proneural-like CD133+ primary patient-derived GBM stem cell (GSC) line that grows as a heterogeneous tumour sphere [29, 35]. R54 cells were grown in serum-free Dulbecco’s modified Eagle medium (DMEM):F12 (1:1; PAN-Biotech, Aidenbach, Germany) containing 1.2 g/l NaHCO3, supplemented with 2% Neuropan 27 (PAN-Biotech), 1% of 200 mM L-Glutamine (Thermo Fisher Scientific, Waltham, USA), 20 ng/ml recombinant human epidermal growth factor (EGF; R&D Systems, Minneapolis, USA), and 20 ng/ml recombinant human fibroblast growth factor (FGF; 154 a.a.; Thermo Fisher Scientific). They were regularly confirmed to be mycoplasma-negative via PCR. Medium with a pH of 6.6 was prepared with powdered medium not containing NaHCO3, which was added at a concentration of 0.4002 g/l, and also contained 15 mM HEPES, additionally to the supplements listed above. Cells were split with 0.05% Trypsin–EDTA solution (Thermo Fisher Scientific) into single cells once per week for regular cell maintenance (37 °C, 5% CO2).
qRT-PCR
R54 GSCs were incubated in pH 7.4 or pH 6.6 for 3 days. RNA was then isolated from GSCs using the NucleoSpin RNA isolation kit (Macherey–Nagel, Düren, Germany), and concentration and quality were measured with the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). RNA was reverse transcribed into cDNA with the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), according to the manufacturers’ instruction. Quantitative real-time PCR (qRT-PCR) was conducted using 20 ng of cDNA template per reaction, as well as the Luna mastermix (NEB) in combination with FAM-MGB labelled hydrolysis (TaqMan™) probes from Thermo Fisher Scientific, namely: ASIC1a (Hs00952802), ASIC1b (Lot 1,431,812), ASIC2 (Hs00153756), ASIC3 (Hs00245092), ASIC4 (Hs00539823), nestin (Hs00707120), GFAP (Hs00909233), TUBB3 (Hs00801390), and GALC (Hs00164660), with GAPDH as a housekeeping gene (Hs02758991). Reactions containing 5 µl Luna NEB mastermix, 1 µl cDNA, 1 µl TaqMan probe, and 3 µl sterile water were pipetted into 4-Strip 0.1 ml Tubes (STARLAB, Hamburg, Germany) and transferred to the Rotor-Gene Q thermocycler (QIAGEN, Hilden, Germany) for real-time measurements. Reactions were performed in technical triplicates for each biological replicate, with technical duplicate negative controls for each TaqMan probe. The qRT-PCR programme was conducted with an initial denaturation step (95 °C, 20 s), 40 cycles of denaturation (95 °C, 30 s), annealing (60 °C, 20 s), and elongation (72 °C, 20 s), followed by a final elongation step (72 °C, 2 min). Experiments were repeated with RNA isolated from at least n = 3 independent cell batches. The Rotor-Gene Q software (Version 1.7.87, QIAGEN) was then used for analysis. Statistical analysis was performed with ΔCt values using an unpaired two-sided Student’s t test, comparing R54 cells at pH 7.4 and pH 6.6, as well as R54 cells and R54 ASIC1 KO cells at pH 7.4. ΔCt values are normalized logarithmic expression values and as such expected to be normally distributed [11].
Sphere migration assay
We used a 2.5D sphere migration assay (SMA) that relies on the attachment of spheres to laminin-coated plates and the subsequent migration of individual GSCs out of the spheres. The day before the experiment, 24-well plates (Thermo Fisher Scientific) were coated with 180 µl laminin from mouse Engelbreth-Holm-Swarm (EHS; Thermo Fisher Scientific) sarcoma (20 ng/µl), resulting in a final laminin density of 1–2 µg per cm2. The plates were then incubated overnight at 4 °C. GSCs were grown for 7 or 14 days into spheres; for some experiments, spheres were grown for 14 days in experimental pH conditions. The spheres were then gently spun down (400 g for 1.5 min) and resuspended in medium with different pH, with and without pharmacological inhibitors. Medium of ASIC2a over-expressing GSCs also contained 4-hydroxytamoxifen (4-HT). For 4-HT induction, the spheres had already been preincubated for 3 days with 4-HT at a concentration of 100 nM. Afterwards, a bystander blinded the experiment. The laminin was aspirated, the wells were washed once with PBS, and then the spheres were gently added into the wells, at a concentration that was empirically determined previously. The 24-well plate was then incubated at 37 °C, 5% CO2 for 5–6 h. Afterwards, the plates were carefully placed under the microscope and pictures taken with the IC Measure programme (The Imaging Source, Version 2.3.1). For experiments conducted under hypoxic conditions, the 24-well plates were incubated for 5–6 h in an Incucyte Live-Cell Analysis System (Sartorius AG, Göttingen, Germany), kindly provided by Dr. Jochen Maurer, at 37 °C, 5% CO2, < 3% O2. Pictures were taken from the Incucyte System with the built-in analysis system. Diameter of spheres and number of migrating cells were counted by hand and positions of cells marked, using ImageJ (NIH, Version 1.52a). Tumourspheres with a high background of freely migrating cells were disregarded, and only cells that could be clearly assigned to a tumoursphere were counted. Furthermore, only spheres with a diameter > 50 µm were quantified. After counting, the experiment was unblinded. The effect of ASICs on migration was tested using PcTx1 (Smartox Biotech, Saint-Egrevè, France), APETx2 (Alomone Labs, Jerusalem, Israel), or MitTx (Smartox Biotech) and the effect of KCNN4 and PI3K by TRAM-34 (Selleck Chemicals, Houston, USA) and wortmannin (Selleck Chemicals), respectively.
Lentiviral transduction
A two-vector inducible system was used to generate ASIC2 overexpressing R54 cells. The cells were transduced first with a lentiviral vector pF GEV16 Super PGKHygro, which expresses a Gal4 DNA binding domain fused to a mutant oestrogen receptor and GEV16. Secondly, the cells were transduced with a lentiviral vector pF 5 × UAS W SV40 Puro ASIC2, which expresses ASIC2 in a Gal4-dependent manner. To generate lentiviral supernatants, 293 T cells were transfected with 3 mg pMD2.G, 5 mg pMDlg/pRRE, and 2.5 mg pRSV-Rev of the lentiviral packaging vectors [41] together with the constructs described above. The supernatants were harvested 48 h post-transfection, filtered (45 mm filter; Schleicher & Schuell, Keene, USA), and were added to R54 cells with 5 mg/ml polybrene, and R54 cells were spin-infected. Stable cell lines were selected in puromycin (1 mg/ml). The expression of ASIC2 was induced by exposure of the cells to 100 nM 4-hydroxytamoxifen for 24 h.
Patch clamp
R54 GSCs overexpressing ASIC2 by lentiviral transduction were induced with 100 nM 4-HT 3 days before splitting. After splitting with 0.05% Trypsin/EDTA (Thermo Fisher Scientific), single cells were seeded on coverslips coated with poly-D-lysine and analysed by patch-clamp during the following 48 h. An Axopatch 200B amplifier and Digidata 1440A digitizer (Molecular Devices, San Jose, USA) controlled by Clampex (Version 10.6) were used for current recordings. The holding potential was − 70 mV. Micropipettes with a resistance of 4–6 MΩ were filled with an intracellular solution containing (in mM) 95 K-gluconate, 30 KCl, 1.2 NaH2PO4, 4.8 Na2HPO4, 5 glucose, 2.38 MgCl2, 1 EGTA, and 0.726 Ca2+-gluconate. The pH was adjusted to pH 7.2 using KOH and HCl. The cells were recorded in a perfused bath system at RT with an extracellular solution containing (in mM) 115 NaCl, 0.4 KH2PO4, 1.6 K2HPO4, 5 Glucose, 1 MgCl2, 25 Na+-gluconate, 3 Ca2+-gluconate, and 5 HEPES/MES. HEPES was used as a buffer for the standard bath solution of pH 7.3, and MES was used as a buffer for the stimulating bath solutions of pH 6 and 5. The pH-values were adjusted using NaOH and HCl. Each stimulation lasted 10 s. The cells were stimulated at least once with pH 6 before being activated with pH 5. Between stimulations, the cells were perfused with standard bath solution pH 7.3 for at least 60 s. To test the effect of PcTx1, 50 nM PcTx1 was added to the pH 6 and the pH 7.3 solution. It was pre-applied for 2 min. For analysis, the software ClampFit was used. Only fast activating (time to peak < 1.5 s) and transient currents with amplitudes > 10 pA were considered as ASIC-currents.
Statistical analysis
Statistical analyses of qRT-PCR results were performed using two-sided t test in Prism 8 (Version 8.4.3), with significance threshold set to p ≤ 0.05.
For sphere migration assays (SMAs), the experimenter was blinded for the condition. We performed SMAs with at least three independent biological replicates with a variable number of technical replicates. To avoid pseudoreplication due to clustering of technical replicates from one biological replicate [28] and to account for the variable number of technical replicates, we statistically analysed SMAs using generalized linear mixed modelling (GLMM). GLMM was performed with RStudio “Prairie Trillium” (Build 461), using the packages glmmADMB [10, 44] and multcomp [19]. We neither saw zero-inflation nor overdispersion in our datasets and determined via Akaike information criterion and chi-square test that negative binomial distribution (migrated cells ~ treatment) provided the best fit as a generalized linear model for SMAs with 7-day old spheres. For SMAs with 14-day-old spheres, which were preconditioned in different pH, a nested model provided the best fit (migrated cells ~ preconditioning pH + preconditioning pH in interaction with assay pH). All models accounted for biological replicates as a random factor. Treatment conditions were then compared with a one-step Tukey’s all-pair comparison, using the glht function of the multcomp package, with significance threshold set to padj < 0.05.
Sphere diameters were analysed with one-way ANOVA in Prism 8 (Version 8.4.3), with significance set at p < 0.05. Normal distribution of the data for one-way ANOVA was confirmed via Anderson–Darling, D’Agostino-Pearson omnibus normality, Shapiro–Wilk normality and Kolmogorov–Smirnov normality tests in Prism 8 (Version 8.4.3), with significance threshold set to p < 0.05.
Availability of data and materials
Datasets generated and analysed during the course of this study are included in this published article. Additional information is available from the corresponding author upon reasonable request.
References
Arcaro A (2016) The role of the PI3K/AKT/mTOR pathway in brain tumor metastasis. J Cancer Metastasis Treat 2:80–89
Babini E, Paukert M, Geisler H-S, Gründer S (2002) Alternative splicing and interaction with di-and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1). J Biol Chem 277:41597–41603
Blaszczak W, Swietach P (2021) What do cellular responses to acidity tell us about cancer? Cancer Metastasis Rev 40:1159–1176
Catacuzzeno L, Fioretti B, Franciolini F (2012) Expression and role of the intermediate-conductance calcium-activated potassium channel KCa3. 1 in glioblastoma. J Signal Transduct 2012:421564. https://doi.org/10.1155/2012/421564
Clusmann J, Cortés Franco K-D, Corredor Suárez D, Katona I, Girbes Mingues M, Boersch N, Pissas K-P, Vanek J, Tian Y, Gründer S (2022) Acidosis induces RIPK1-dependent death of glioblastoma stem cells via acid-sensing ion channel 1a. Cell Death Dis
d’Alessandro G, Catalano M, Sciaccaluga M, Chece G, Cipriani R, Rosito M, Grimaldi A, Lauro C, Cantore G, Santoro A (2013) KCa3. 1 channels are involved in the infiltrative behavior of glioblastoma in vivo. Cell Death Dis 4:e773–e773
Damaghi M, Wojtkowiak JW, Gillies RJ (2013) pH sensing and regulation in cancer. Front Physiol 4:370
Ding M, Zhang S, Guo Y, Yao J, Shen Q, Huang M, Chen W, Yu S, Zheng Y, Lin Y (2022) Tumor microenvironment acidity triggers lipid accumulation in liver cancer via SCD1 activation. Mol Cancer Res 20:810–822
Esencay M, Sarfraz Y, Zagzag D (2013) CXCR7 is induced by hypoxia and mediates glioma cell migration towards SDF-1α. BMC Cancer 13:1–9
Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder MN, Nielsen A, Sibert J (2012) AD model builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim Methods Softw 27:233–249. https://doi.org/10.1080/10556788.2011.597854
Friedman N, Cai L, Xie XS (2006) Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett 97:168302
Gage FH, Ray J, Fisher LJ (1995) Isolation, characterization, and use of stem cells from the CNS. Annu Rev Neurosci 18:159–192
Gerweck LE, Seetharaman K (1996) Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res 56:1194–1198
Gravendeel LAM, Kouwenhoven MCM, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LBC, Kloosterhof NK (2009) Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res 69:9065–9072
Grifoni SC, Jernigan NL, Hamilton G, Drummond HA (2008) ASIC proteins regulate smooth muscle cell migration. Microvasc Res 75:202–210. https://doi.org/10.1016/j.mvr.2007.08.003
Grimaldi A, D’alessandro G, Golia MT, Grössinger EM, Di Angelantonio S, Ragozzino D, Santoro A, Esposito V, Wulff H, Catalano M (2016) KCa3. 1 inhibition switches the phenotype of glioma-infiltrating microglia/macrophages. Cell Death Dis 7:e2174–e2174
Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU (2017) Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac J Cancer Prev APJCP 18:3
Hessmann E, Buchholz SM, Demir IE, Singh SK, Gress TM, Ellenrieder V, Neesse A (2020) Microenvironmental determinants of pancreatic cancer. Physiol Rev 100:1707–1751
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biometrical J 50:346–363. https://doi.org/10.1002/bimj.200810425
Huang W, Ding X, Ye H, Wang J, Shao J, Huang T (2018) Hypoxia enhances the migration and invasion of human glioblastoma U87 cells through PI3K/Akt/mTOR/HIF-1α pathway. NeuroReport 29:1578–1585
Jiang N, Dai Q, Su X, Fu J, Feng X, Peng J (2020) Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol Biol Rep 47:4587–4629
Joeres N, Augustinowski K, Neuhof A, Assmann M, Gründer S (2016) Functional and pharmacological characterization of two different ASIC1a/2a heteromers reveals their sensitivity to the spider toxin PcTx1. Sci Rep 6:1–14
Joseph JV, Conroy S, Pavlov K, Sontakke P, Tomar T, Eggens-Meijer E, Balasubramaniyan V, Wagemakers M, den Dunnen WFA, Kruyt FAE (2015) Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1α–ZEB1 axis. Cancer Lett 359:107–116
Kapoor N, Bartoszewski R, Qadri YJ, Bebok Z, Bublen JK, Fuller CM, Benos DJ (2009) Knockdown of ASIC1 and epithelial sodium channel subunits inhibits glioblastoma whole cell current and cell migration. J Biol Chem 284:24526–24541. https://doi.org/10.1074/jbc.M109.037390
Kaushal V, Koeberle PD, Wang Y, Schlichter LC (2007) The Ca2+-activated K+ channel KCNN4/KCa3. 1 contributes to microglia activation and nitric oxide-dependent neurodegeneration. J Neurosci 27:234–244
Klumpp L, Sezgin EC, Skardelly M, Eckert F, Huber SM (2018) KCa3. 1 channels and glioblastoma: in vitro studies. Curr Neuropharmacol 16:627–635
Lai W, Liu L, Zeng Y, Wu H, Xu H, Chen S, Chu Z (2013) KCNN4 Channels participate in the EMT induced by PRL-3 in colorectal cancer. Med Oncol 30:566. https://doi.org/10.1007/s12032-013-0566-z
Lazic SE, Clarke-Williams CJ, Munafò MR (2018) What exactly is ‘N’in cell culture and animal experiments? PLoS Biol 16:e2005282
Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9:391–403
Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA (2008) Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res 6:21–30
Li Q-T, Feng Y-M, Ke Z-H, Qiu M-J, He X-X, Wang M-M, Li Y-N, Xu J, Shi L-L, Xiong Z-F (2019) KCNN4 promotes invasion and metastasis through the MAPK/ERK pathway in hepatocellular carcinoma. J Investig Med 68:68–74. https://doi.org/10.1136/jim-2019-001073
Li X, Wu C, Chen N, Gu H, Yen A, Cao L, Wang E, Wang L (2016) PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 7:33440
Liu Y, Zhao L, Ma W, Cao X, Chen H, Feng D, Liang J, Yin K, Jiang X (2015) The blockage of KCa3. 1 channel inhibited proliferation, migration and promoted apoptosis of human hepatocellular carcinoma cells. J Cancer 6:643
Lötsch D, Steiner E, Holzmann K, Spiegl-Kreinecker S, Pirker C, Hlavaty J, Petznek H, Hegedus B, Garay T, Mohr T (2013) Major vault protein supports glioblastoma survival and migration by upregulating the EGFR/PI3K signalling axis. Oncotarget 4:1904
Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J (2010) Transcriptional profiles of CD133+ and CD133− glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res 70:2030–2040
Martínez D, Vermeulen M, Trevani A, Ceballos A, Sabatté J, Gamberale R, Álvarez ME, Salamone G, Tanos T, Coso OA (2006) Extracellular acidosis induces neutrophil activation by a mechanism dependent on activation of phosphatidylinositol 3-kinase/Akt and ERK pathways. J Immunol 176:1163–1171
Oh J, Sahgal A, Sanghera P, Tsao MN, Davey P, Lam K, Symons S, Aviv R, Perry JR (2011) Glioblastoma: patterns of recurrence and efficacy of salvage treatments. Can J Neurol Sci 38:621–625
Potter M, Newport E, Morten KJ (2016) The Warburg effect: 80 years on. Biochem Soc Trans 44:1499–1505
Qiao X, Xu J, Yang Q-J, Du Y, Lei S, Liu Z-H, Liu X, Liu H (2013) Transient acidosis during early reperfusion attenuates myocardium ischemia reperfusion injury via PI3k-Akt-eNOS signaling pathway. Oxid Med Cell Longev 2013:126083. https://doi.org/10.1155/2013/126083
Rooj AK, McNicholas CM, Bartoszewski R, Bebok Z, Benos DJ, Fuller CM (2012) Glioma-specific cation conductance regulates migration and cell cycle progression. J Biol Chem 287:4053–4065. https://doi.org/10.1074/jbc.M111.311688
Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33:401–406
Ruggieri P, Mangino G, Fioretti B, Catacuzzeno L, Puca R, Ponti D, Miscusi M, Franciolini F, Ragona G, Calogero A (2012) The inhibition of KCa3. 1 channels activity reduces cell motility in glioblastoma derived cancer stem cells. Plos One 7:e47825. https://doi.org/10.1371/journal.pone.0047825
Sheng Y, Wu B, Leng T, Zhu L, Xiong Z (2021) Acid-sensing ion channel 1 (ASIC1) mediates weak acid-induced migration of human malignant glioma cells. Am J Cancer Res 11:997
Skaug H, Fournier D, Bolker B, Magnusson A, Nielsen A (2016) Generalized linear mixed models using AD model builder. R Package version 08. https://r-forge.r-project.org/projects/glmmadmb/
Stock C, Schwab A (2009) Protons make tumor cells move like clockwork. Pflügers Arch J Physiol 458:981–992
Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996
Tian Y, Bresenitz P, Reska A, El Moussaoui L, Beier CP, Gründer S (2017) Glioblastoma cancer stem cell lines express functional acid sensing ion channels ASIC1a and ASIC3. Sci Rep 7:1–14
Turner KL, Honasoge A, Robert SM, McFerrin MM, Sontheimer H (2014) A proinvasive role for the Ca2+-activated K+ channel KCa3. 1 in malignant glioma. Glia 62:971–981
Vila-Carriles WH, Kovacs GG, Jovov B, Zhou Z-H, Pahwa AK, Colby G, Esimai O, Gillespie GY, Mapstone TB, Markert JM (2006) Surface expression of ASIC2 inhibits the amiloride-sensitive current and migration of glioma cells. J Biol Chem 281:19220–19232
Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M (1997) A proton-gated cation channel involved in acid-sensing. Nature 386:173–177
Webb BA, Chimenti M, Jacobson MP, Barber DL (2011) Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer 11:671–677
Wu J, Xu Y, Jiang Y-Q, Xu J-P, Hu Y, Zha X (2016) ASIC subunit ratio and differential surface trafficking in the brain. Mol Brain 9:4. https://doi.org/10.1186/s13041-016-0185-7
Wu W, Klockow JL, Zhang M, Lafortune F, Chang E, Jin L, Wu Y, Daldrup-Link HE (2021) Glioblastoma multiforme (GBM): an overview of current therapies and mechanisms of resistance. Pharmacol Res 171:105780
Xu P, Mo X, Xia R, Jiang L, Zhang C, Xu H, Sun Q, Zhou G, Zhang Y, Wang Y, Xia H (2021) KCNN4 promotes the progression of lung adenocarcinoma by activating the AKT and ERK signaling pathways. Cancer Biomark 31:1–15. https://doi.org/10.3233/CBM-201045
Zhang Z, Yao L, Yang J, Wang Z, Du G (2018) PI3K/Akt and HIF-1 signaling pathway in hypoxia-ischemia. Mol Med Rep 18:3547–3554
Zhao H, Wang J, Shao W, Wu C, Chen Z, To ST, Li W (2017) Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: current preclinical and clinical development. Mol Cancer 16:1–16
Zhou Z, Song J, Li W, Liu X, Cao L, Wan L, Tan Y, Ji S, Liang Y, Gong F (2017) The acid-sensing ion channel, ASIC2, promotes invasion and metastasis of colorectal cancer under acidosis by activating the calcineurin/NFAT1 axis. J Exp Clin Cancer Res 36:1–12
Zhu S, Zhou H-Y, Deng S-C, Deng S-J, He C, Li X, Chen J-Y, Jin Y, Hu Z-L, Wang F (2017) ASIC1 and ASIC3 contribute to acidity-induced EMT of pancreatic cancer through activating Ca2+/RhoA pathway. Cell Death Dis 8:e2806–e2806
Acknowledgements
We thank J. Maurer, Aachen, for helpful discussions and for providing access to the Incucyte System and J. Clusmann, Aachen, for comments on the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by grant 124/18 of the START programme of the Faculty of Medicine at RWTH Aachen University to YT.
Author information
Authors and Affiliations
Contributions
K-DCF performed migration assays and qPCR. ICB performed patch-clamp experiments. MAF conducted the lentiviral transduction. K-DCF performed the statistical analysis. K-DCF and ICB generated figures. K-DCF, YT, and SG designed the study. DPD, YT, and SG supervised the study. K-DCF and SG wrote the manuscript with important input from all other authors.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Conflict of interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cortés Franco, KD., Brakmann, I.C., Feoktistova, M. et al. Aggressive migration in acidic pH of a glioblastoma cancer stem cell line in vitro is independent of ASIC and KCa3.1 ion channels, but involves phosphoinositide 3-kinase. Pflugers Arch - Eur J Physiol 475, 405–416 (2023). https://doi.org/10.1007/s00424-022-02781-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-022-02781-w