Abstract
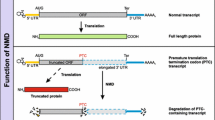
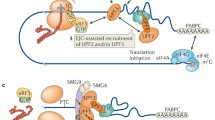
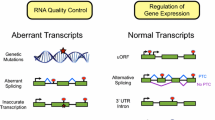
Nonsense-mediated messenger RNA (mRNA) decay (NMD) is a quality control mechanism that degrades irregular or faulty mRNAs. NMD mainly degrades mRNAs, which contain a premature termination codon (PTC) and therefore encode a truncated protein. Furthermore, NMD alters the expression of different types of cellular mRNAs, the so-called endogenous NMD substrates. In this review, we focus on the impact of NMD on cellular and molecular physiology. We specify key classes of NMD substrates and provide a detailed overview of the physiological function of gene regulation by NMD. We also describe different mechanisms of NMD substrate degradation and how the regulation of the NMD machinery affects cellular physiology. Finally, we outline the physiological functions of central NMD factors.






Similar content being viewed by others
References
Abernathy E, Glaunsinger B (2015) Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 479–480:600–608. doi:10.1016/j.virol.2015.02.007
Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Inoff-Germain G, Gochman P, Long R, Rapoport JL, Rouleau GA (2011) A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry 16:238–239. doi:10.1038/mp.2010.59
Ahlquist P (2006) Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol 4:371–382. doi:10.1038/nrmicro1389
Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ (2008) Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14:914–927. doi:10.1261/rna.829208
Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL (2004) Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood 103:1876–1882. doi:10.1182/blood-2003-06-1859
Anko ML, Neugebauer KM (2012) RNA-protein interactions in vivo: global gets specific. Trends Biochem Sci 37:255–262. doi:10.1016/j.tibs.2012.02.005
Applequist SE, Selg M, Raman C, Jack HM (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25:814–821
Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16:433–439. doi:10.1016/j.cub.2006.01.018
Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S (2008) MicroRNA expression in the adult mouse central nervous system. RNA 14:432–444. doi:10.1261/rna.783108
Balistreri G, Horvath P, Schweingruber C, Zund D, McInerney G, Merits A, Muhlemann O, Azzalin C, Helenius A (2014) The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe 16:403–411. doi:10.1016/j.chom.2014.08.007
Barker GF, Beemon K (1994) Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol Cell Biol 14:1986–1996
Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24:3470–3481. doi:10.1038/sj.emboj.7600777
Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336. doi:10.1146/annurev.biochem.72.121801.161720
Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, Wouters BG, Bell JC (2004) Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 24:7469–7482. doi:10.1128/MCB.24.17.7469-7482.2004
Boehm V, Haberman N, Ottens F, Ule J, Gehring NH (2014) 3′ UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep 9:555–568. doi:10.1016/j.celrep.2014.09.012
Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL (2007) A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21:1636–1652. doi:10.1101/gad.1558107
Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT (2004) The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell 14:585–598. doi:10.1016/j.molcel.2004.05.005
Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF (2011) Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 42:500–510. doi:10.1016/j.molcel.2011.04.018
Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E (2010) Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci U S A 107:10050–10055. doi:10.1073/pnas.1000993107
Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270:28995–29003
Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Spencer TA, Krause BR, Erickson SK, Farese RV Jr (1998) ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J Biol Chem 273:26755–26764
Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF (2009) A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 16:747–753. doi:10.1038/nsmb.1612
Chen Z, Gore BB, Long H, Ma L, Tessier-Lavigne M (2008) Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron 58:325–332. doi:10.1016/j.neuron.2008.02.016
Clerici M, Deniaud A, Boehm V, Gehring NH, Schaffitzel C, Cusack S (2014) Structural and functional analysis of the three MIF4G domains of nonsense-mediated decay factor UPF2. Nucleic Acids Res 42:2673–2686. doi:10.1093/nar/gkt1197
Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S (2009) Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28:2293–2306. doi:10.1038/emboj.2009.175
Colak D, Ji SJ, Porse BT, Jaffrey SR (2013) Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 153:1252–1265. doi:10.1016/j.cell.2013.04.056
Desbois C, Rousset R, Bantignies F, Jalinot P (1996) Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science 273:951–953
Donnelly N, Gorman AM, Gupta S, Samali A (2013) The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 70:3493–3511. doi:10.1007/s00018-012-1252-6
Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16:49–55. doi:10.1038/nsmb.1530
Garcia D, Garcia S, Voinnet O (2014) Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16:391–402. doi:10.1016/j.chom.2014.08.001
Gardner LB (2008) Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol 28:3729–3741. doi:10.1128/MCB.02284-07
Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW (2009) Disassembly of exon junction complexes by PYM. Cell 137:536–548. doi:10.1016/j.cell.2009.02.042
Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA (2012) BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol 22:2135–2139. doi:10.1016/j.cub.2012.09.020
Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20:881–890. doi:10.1016/j.molcel.2005.10.026
Graveley BR (2000) Sorting out the complexity of SR protein functions. RNA 6:1197–1211
Hamid FM, Makeyev EV (2014) Regulation of mRNA abundance by polypyrimidine tract-binding protein-controlled alternate 5′ splice site choice. PLoS Genet 10, e1004771. doi:10.1371/journal.pgen.1004771
Harding CL, Lloyd DR, McFarlane CM, Al-Rubeai M (2000) Using the Microcyte flow cytometer to monitor cell number, viability, and apoptosis in mammalian cell culture. Biotechnol Prog 16:800–802. doi:10.1021/bp0000813
Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11:619–633
Hirschfeld M, zur Hausen A, Bettendorf H, Jager M, Stickeler E (2009) Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer Res 69:2082–2090. doi:10.1158/0008-5472.CAN-08-1997
Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14:2609–2617. doi:10.1261/rna.1386208
Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27:736–747. doi:10.1038/emboj.2008.17
Kadlec J, Izaurralde E, Cusack S (2004) The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11:330–337. doi:10.1038/nsmb741
Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF (2015) The unfolded protein response is shaped by the NMD pathway. EMBO Rep 16:599–609. doi:10.15252/embr.201439696
Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20:355–367. doi:10.1101/gad.1389006
Keppetipola N, Sharma S, Li Q, Black DL (2012) Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol 47:360–378. doi:10.3109/10409238.2012.691456
Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11:345–355. doi:10.1038/nrg2776
Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH (2006) Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12:1015–1022. doi:10.1261/rna.12506
Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE (2007) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446:926–929. doi:10.1038/nature05676
Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, Briault S, Fryns JP, Hamel B, Chelly J, Ropers HH, Ronce N, Blesson S, Moraine C, Gecz J, Raynaud M (2010) Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry 15:767–776. doi:10.1038/mp.2009.14
Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001) The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20:4987–4997. doi:10.1093/emboj/20.17.4987
Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19:6860–6869. doi:10.1093/emboj/19.24.6860
Lewis BP, Green RE, Brenner SE (2003) Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A 100:189–192. doi:10.1073/pnas.0136770100
Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T, Chen YS, Groth M, Krueger A, Platzer M, Yang YG, Rudolph KL, Wang ZQ (2015) Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. doi:10.15252/embj.201489947
Li Z, Nagy PD (2011) Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol 8:305–315
Liu C, Karam R, Zhou Y, Su F, Ji Y, Li G, Xu G, Lu L, Wang C, Song M, Zhu J, Wang Y, Zhao Y, Foo WC, Zuo M, Valasek MA, Javle M, Wilkinson MF, Lu Y (2014) The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med 20:596–598. doi:10.1038/nm.3548
Loh B, Jonas S, Izaurralde E (2013) The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27:2125–2138. doi:10.1101/gad.226951.113
Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M (2004) Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 42:213–223
Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF (2014) Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 6:748–764. doi:10.1016/j.celrep.2014.01.028
Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121–1131
Lykke-Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, Sandelin A, Jensen TH (2014) Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev 28:2498–2517. doi:10.1101/gad.246538.114
Lynch SA, Nguyen LS, Ng LY, Waldron M, McDonald D, Gecz J (2012) Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet 55:476–479. doi:10.1016/j.ejmg.2012.03.010
Makeyev EV, Zhang J, Carrasco MA, Maniatis T (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27:435–448. doi:10.1016/j.molcel.2007.07.015
Matlin AJ, Clark F, Smith CW (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6:386–398. doi:10.1038/nrm1645
McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW (2010) Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A 107:12186–12191. doi:10.1073/pnas.1007336107
Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10:99–105
Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol 20:8944–8957
Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36:1073–1078. doi:10.1038/ng1429
Menendez JA, Mehmi I, Griggs DW, Lupu R (2003) The angiogenic factor CYR61 in breast cancer: molecular pathology and therapeutic perspectives. Endocr Relat Cancer 10:141–152
Mocquet V, Neusiedler J, Rende F, Cluet D, Robin JP, Terme JM, Duc Dodon M, Wittmann J, Morris C, Le Hir H, Ciminale V, Jalinot P (2012) The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol 86:7530–7543. doi:10.1128/JVI.07021-11
Morris-Desbois C, Bochard V, Reynaud C, Jalinot P (1999) Interaction between the Ret finger protein and the Int-6 gene product and co-localisation into nuclear bodies. J Cell Sci 112(Pt 19):3331–3342
Morrison M, Harris KS, Roth MB (1997) smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci U S A 94:9782–9785
Mort M, Ivanov D, Cooper DN, Chuzhanova NA (2008) A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29:1037–1047. doi:10.1002/humu.20763
Muhlrad D, Parker R (1999) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299–1307
Nagy E, Maquat LE (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23:198–199
Nakano K, Ando T, Yamagishi M, Yokoyama K, Ishida T, Ohsugi T, Tanaka Y, Brighty DW, Watanabe T (2013) Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect 15:491–505. doi:10.1016/j.micinf.2013.03.006
Narayanan K, Makino S (2013) Interplay between viruses and host mRNA degradation. Biochim Biophys Acta 1829:732–741. doi:10.1016/j.bbagrm.2012.12.003
Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21:708–718. doi:10.1101/gad.1525507
O’Donnell M, Chance RK, Bashaw GJ (2009) Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci 32:383–412. doi:10.1146/annurev.neuro.051508.135614
Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S (2012) N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40:1251–1266. doi:10.1093/nar/gkr791
Oren YS, McClure ML, Rowe SM, Sorscher EJ, Bester AC, Manor M, Kerem E, Rivlin J, Zahdeh F, Mann M, Geiger T, Kerem B (2014) The unfolded protein response affects readthrough of premature termination codons. EMBO Mol Med 6:685–701. doi:10.1002/emmm.201303347
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415. doi:10.1038/ng.259
Pan Y, Chen H, Siu F, Kilberg MS (2003) Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem 278:38402–38412. doi:10.1074/jbc.M304574200
Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ Jr, Dietz HC (1996) Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci U S A 93:10928–10932
Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev 2:215–227
Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9:265–273. doi:10.1038/nrc2620
Rahman L, Bliskovski V, Kaye FJ, Zajac-Kaye M (2004) Evolutionary conservation of a 2-kb intronic sequence flanking a tissue-specific alternative exon in the PTBP2 gene. Genomics 83:76–84
Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, Ott M (2015) A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57:329–340. doi:10.1016/j.molcel.2014.12.028
Rigby RE, Rehwinkel J (2015) RNA degradation in antiviral immunity and autoimmunity. Trends Immunol 36:179–188. doi:10.1016/j.it.2015.02.001
Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG (2010) The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 120:127–141. doi:10.1172/JCI40027
Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ (2008) Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28:4320–4330. doi:10.1128/MCB.00361-08
Schmidt SA, Foley PL, Jeong DH, Rymarquis LA, Doyle F, Tenenbaum SA, Belasco JG, Green PJ (2015) Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res 43:309–323. doi:10.1093/nar/gku1258
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5:R13. doi:10.1186/gb-2004-5-3-r13
Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M (1994) The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell 78:409–424
Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21:209–223. doi:10.1128/MCB.21.1.209-223.2001
Seward SL Jr, Gahl WA (2013) Hermansky-Pudlak syndrome: health care throughout life. Pediatrics 132:153–160. doi:10.1542/peds.2012-4003
Shepard PJ, Hertel KJ (2009) The SR protein family. Genome Biol 10:242. doi:10.1186/gb-2009-10-10-242
Sheth U, Parker R (2006) Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095–1109. doi:10.1016/j.cell.2006.04.037
Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK, Lou CH, Espinoza JL, Song HW, Phan MH, Ramaiah M, Huang L, McCarrey JR, Peterson KJ, De Rooij DG, Cook-Andersen H, Wilkinson MF (2016) The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell. doi:10.1016/j.cell.2016.02.046
Silvain J, Collet JP, Nagaswami C, Beygui F, Edmondson KE, Bellemain-Appaix A, Cayla G, Pena A, Brugier D, Barthelemy O, Montalescot G, Weisel JW (2011) Composition of coronary thrombus in acute myocardial infarction. J Am Coll Cardiol 57:1359–1367. doi:10.1016/j.jacc.2010.09.077
Simons-Evelyn M, Young HA, Anderson SK (1997) Characterization of the mouse Nktr gene and promoter. Genomics 40:94–100. doi:10.1006/geno.1996.4562
Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ (2012) The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151:750–764. doi:10.1016/j.cell.2012.10.007
Singh G, Rebbapragada I, Lykke-Andersen J (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6, e111. doi:10.1371/journal.pbio.0060111
Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG (2005) Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21:1469–1477. doi:10.1111/j.1460-9568.2005.03978.x
Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi:10.1016/j.cell.2009.01.042
Spellman R, Llorian M, Smith CW (2007) Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27:420–434. doi:10.1016/j.molcel.2007.06.016
Stockklausner C, Breit S, Neu-Yilik G, Echner N, Hentze MW, Kulozik AE, Gehring NH (2006) The uORF-containing thrombopoietin mRNA escapes nonsense-mediated decay (NMD). Nucleic Acids Res 34:2355–2363. doi:10.1093/nar/gkl277
Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J (2001) SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20:1785–1796. doi:10.1093/emboj/20.7.1785
Susova O, Gurtsevich VE (2003) The role of region pX in the life cycle of HTLV-I and in carcinogenesis. Mol Biol 37:392–403
Talloczy Z, Jiang W, Virgin HW, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B (2002) Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A 99:190–195. doi:10.1073/pnas.012485299
Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N (2012) Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9:1370–1379. doi:10.4161/rna.22360
Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O’Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J, Varian J, West S, Widaa S, Teague J, Dicks E, Butler A, Menzies A, Richardson D, Jenkinson A, Shepherd R, Raine K, Moon J, Luo Y, Parnau J, Bhat SS, Gardner A, Corbett M, Brooks D, Thomas P, Parkinson-Lawrence E, Porteous ME, Warner JP, Sanderson T, Pearson P, Simensen RJ, Skinner C, Hoganson G, Superneau D, Wooster R, Bobrow M, Turner G, Stevenson RE, Schwartz CE, Futreal PA, Srivastava AK, Stratton MR, Gecz J (2007) Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39:1127–1133. doi:10.1038/ng2100
Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7:131–142. doi:10.1038/nrm1835
Trusolino L, Comoglio PM (2002) Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nat Rev Cancer 2:289–300. doi:10.1038/nrc779
Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G (2010) Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol 191:87–99. doi:10.1083/jcb.201001073
Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101:11269–11274. doi:10.1073/pnas.0400541101
Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE (2007) The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res 35:4542–4551. doi:10.1093/nar/gkm461
Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086. doi:10.1126/science.1209038
Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB (2011) Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol 31:3670–3680. doi:10.1128/MCB.05704-11
Wei ML (2006) Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res 19:19–42. doi:10.1111/j.1600-0749.2005.00289.x
Weil JE, Beemon KL (2006) A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 12:102–110. doi:10.1261/rna.2129806
Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT (2008) NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22:1381–1396. doi:10.1101/gad.468808
Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT (2012) Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol 13:R35. doi:10.1186/gb-2012-13-5-r35
Wek RC, Cavener DR (2007) Translational control and the unfolded protein response. Antioxid Redox Signal 9:2357–2371. doi:10.1089/ars.2007.1764
Wengrod J, Martin L, Wang D, Frischmeyer-Guerrerio P, Dietz HC, Gardner LB (2013) Inhibition of nonsense-mediated RNA decay activates autophagy. Mol Cell Biol 33:2128–2135. doi:10.1128/MCB.00174-13
Withers JB, Beemon KL (2010) Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon. Retrovirology 7:65. doi:10.1186/1742-4690-7-65
Wittmann J, Hol EM, Jack HM (2006) hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26:1272–1287. doi:10.1128/MCB.26.4.1272-1287.2006
Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW (2004) Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13:91–100
Xu X, Zhang L, Tong P, Xun G, Su W, Xiong Z, Zhu T, Zheng Y, Luo S, Pan Y, Xia K, Hu Z (2013) Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet 83:560–564. doi:10.1111/cge.12014
Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C (2010) The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29:2082–2096. doi:10.1038/emboj.2010.81
Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O (2011) Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17:2108–2118. doi:10.1261/rna.030247.111
Younis I, Green PL (2005) The human T-cell leukemia virus Rex protein. Front Biosci 10:431–445
Zahler AM, Lane WS, Stolk JA, Roth MB (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 6:837–847
Acknowledgments
This research was funded by a grant from the Deutsche Forschungsgemeinschaft (GE2014/4-1) to N.H.G.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Ottens, F., Gehring, N.H. Physiological and pathophysiological role of nonsense-mediated mRNA decay. Pflugers Arch - Eur J Physiol 468, 1013–1028 (2016). https://doi.org/10.1007/s00424-016-1826-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-016-1826-5