Abstract
GABAergic neurons in the neocortex are diverse with regard to morphology, physiology, and axonal targeting pattern, indicating functional specializations within the cortical microcircuitry. Little information is available, however, about functional properties of distinct subtypes of GABAergic neurons in the intact brain. Here, we combined in vivo two-photon calcium imaging in supragranular layers of the mouse neocortex with post hoc immunohistochemistry against the three calcium-binding proteins parvalbumin, calretinin, and calbindin in order to assign subtype marker profiles to neuronal activity. Following coronal sectioning of fixed brains, we matched cells in corresponding volumes of image stacks acquired in vivo and in fixed brain slices. In GAD67-GFP mice, more than 95% of the GABAergic cells could be unambiguously matched, even in large volumes comprising more than a thousand interneurons. Triple immunostaining revealed a depth-dependent distribution of interneuron subtypes with increasing abundance of PV-positive neurons with depth. Most importantly, the triple-labeling approach was compatible with previous in vivo calcium imaging following bulk loading of Oregon Green 488 BAPTA-1, which allowed us to classify spontaneous calcium transients recorded in vivo according to the neurochemically defined GABAergic subtypes. Moreover, we demonstrate that post hoc immunostaining can also be applied to wild-type mice expressing the genetically encoded calcium indicator Yellow Cameleon 3.60 in cortical neurons. Our approach is a general and flexible method to distinguish GABAergic subtypes in cell populations previously imaged in the living animal. It should thus facilitate dissecting the functional roles of these subtypes in neural circuitry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cortical information processing relies on the intricate balance of excitation and inhibition in neocortical networks [33], with inhibition playing a crucial role not only for the control of excitatory signal flow but also for synchronization of neural activity and the generation of rhythmic oscillations [2, 75]. Inhibition in the neocortex is provided by GABAergic interneurons, which constitute around 10–20% of cortical cells and exhibit a remarkable diversity in terms of their morphological characteristics (in particular axonal targeting patterns), electrophysiological properties, and molecular composition [8, 20, 22, 54, 66]. Based on these features, many subtypes of GABAergic interneurons have been defined; however, no consensus on a full catalogue of distinct classes has been reached yet [5, 22]. In addition, little information is available on the in vivo activation patterns of particular subtypes, especially during specific behaviors. This lack of information is due to the fact that GABAergic interneurons are sparsely distributed within cortical microcircuits, which makes it difficult to identify and target these cells in vivo.
Direct studies of behavior-related membrane potential dynamics in specific GABAergic subtypes have become possible only recently with the availability of mice expressing fluorescent proteins in genetically defined neuronal subtypes [56, 61, 68] and employing whole-cell or cell-attached recordings from cells targeted with two-photon microscopy [45]. These studies highlighted the functional diversity of the GABAergic population [16, 26, 51]. In addition to electrophysiological recordings, in vivo two-photon calcium imaging enables activity measurements in neocortical populations (reviewed in [30, 35, 74]). To distinguish excitatory and inhibitory interneurons, a small number of studies have applied bulk loading of synthetic calcium indicators to transgenic mice in which all GABAergic neurons express a fluorescent protein [68], revealing for example differences in orientation tuning in the visual cortex [42, 65]. Further discrimination of GABAergic subtypes is desirable and in principle can be achieved with two methods. First, in vivo calcium imaging can be performed in mice expressing a fluorescent protein in a particular subset of GABAergic neurons. This promising approach has been taken up only very recently [63, 80]. Second, various subtypes of GABAergic neurons may be discriminated post mortem by means of immunohistochemistry (IHC). For that purpose, cells previously imaged in vivo have to be identified in fixed brain slices treated with antibodies against specific chemical markers. The feasibility of such post hoc IHC for discriminating GABAergic subtypes has been recently demonstrated in a few studies [42, 60, 74]. This approach is, however, just emerging, and experimental procedures are not yet well established. Advancing the combination of in vivo calcium imaging and post hoc IHC therefore is an important direction for further dissection of neocortical population activity into its different cellular components.
Here, we present post hoc IHC as a valuable method to identify GABAergic subtypes in cortical networks, from which calcium indicator measurements had been performed in the living mouse. We evaluate the method in terms of post hoc cell identification yield, different antibody stains against calcium-binding proteins, and its applicability to tissue labeled with either synthetic or genetically encoded calcium indicators. We demonstrate spontaneous calcium transients in interneurons of identified subtype. Combining in vivo calcium imaging with post hoc IHC opens numerous opportunities for investigating specific activity patterns within local neural circuits.
Materials and methods
Animal preparation and fluorescence labeling
Anesthesia of both C57BL/6 wild-type and GAD67-GFP (Δneo) mice (2–3 months old, heterozygous, C57BL/6 background; phenotype selected based on transcranial fluorescence of pups) [68] was induced with 5% isoflurane in air, followed by urethane (1.4 mg/g body weight, 20% aqueous solution injected i.p.). During surgery, the level of anesthesia was monitored based on whisker movement and foot-pinch reflex. If required, urethane anesthesia was temporarily supplemented with isoflurane. We performed a craniotomy above the left barrel cortex (centered 3.5 mm lateral, 1 mm post bregma), removed the dura, and superfused the exposed brain with normal rat ringer (NRR) solution (in mM: 145 NaCl, 5.4 KCl, 10 HEPES, 1.0 MgCl2, 1.8 CaCl2; pH 7.2 adjusted with NaOH). For functional calcium measurements, we labeled layer 2/3 (L2/3) cell populations with the green fluorescent calcium indicator Oregon Green 488 BAPTA-1 (OGB-1) by multi-cell bolus loading [25, 67]. Briefly, the acetoxymethyl ester of OGB-1 (OGB-1 AM; Invitrogen, Basel, Switzerland) was dissolved in a solution of 20% Pluronic F-127 in DMSO (both from Sigma-Aldrich, Buchs, Switzerland) and diluted in Ca2+-free NRR to a final concentration of 1 mM. Dye solution was pressure-injected with a patch pipette (300 mbar during 1–2 min) into a barrel column previously identified by intrinsic optical imaging. Selective astrocyte staining was achieved by exposing the brain to 50 μM sulforhodamine 101 (SR101; Sigma-Aldrich, Buchs, Switzerland) during 5–10 min [59]. To dampen heartbeat- and breathing-induced motion artifacts, the cranial window was covered with a layer of agarose (0.5–1 mm; type III-A, Sigma; 1.5% in NRR) and an immobilized cover slip.
Virus-mediated expression of Yellow Cameleon YC3.60
Recombinant adeno-associated virus harboring YC3.60 under control of the human synapsin promoter was produced and purified according to [49] (virus titer in transducing units, 3·108 μL−1). Virus solution was diluted 1:1 with a solution of 20% mannitol in phosphate-buffered saline (PBS; 0.1 M, pH 7.3) and delivered into cortical tissue (volume 200–300 nL; depth about 250 μm) through a glass pipette (tip size 8–12 μm) by stereotactic injection [49]. Animals were imaged 3.5 to 9 months after infection.
Intrinsic optical signal imaging
We employed intrinsic optical signal imaging to identify the position of the cortical representation of a specific whisker in the mouse barrel cortex. Image acquisition was performed with a 12 bit b/w CCD camera (Teli CS3960DCL) and a ×4 objective (UPlanFL N, ×4/0.13, Olympus). Prior to intrinsic imaging, a blood vessel reference map was acquired under green light illumination (peak wavelength 525 nm, Sloan L5-G61N-GT LED). Intrinsic optical imaging was performed under red light illumination (peak wavelength 660 nm, Kingbright L-7113SRD LED) using the HelioScan imaging software package [47]. Briefly, baseline images were acquired during 5 s without stimulation. During the following 5.5 s, a single whisker was tapped (50 Hz rectangular motion along whisker axis using a pipette tip mounted on a loudspeaker), and images were acquired during 5 s after an initial 0.5-s delay. Images from periods without and with stimulation were separately averaged and subtracted. The procedure was repeated at intervals of 20 s until a clear intrinsic signal was obtained.
In vivo two-photon microscopy
We used a custom-built two-photon microscope controlled by the HelioScan imaging software package [47] and powered by a Ti:Sapphire laser system tunable between 710 and 1,090 nm (MaiTai Broadband, Spectra-Physics, Santa Clara, CA, USA). Beam size was adjusted with a telescope and laser intensity modulated with a Pockels cell (model 350/80, controller model 302RM, Conoptics, Danbury, CT, USA). Two galvanometric mirrors (model 6210; Cambridge Technology, Lexington, MA) were used for x/y scanning and a piezoelectric focusing device (P-725.4CD PIFOC, Physik Instrumente, Karlsruhe, Germany) for stack acquisition. Fluorescence signals were acquired with a custom-built three-channel detector system with two dichroic mirrors (DC-Blue and DC-Red from Linos, Göttingen, Germany) (Fig. 1). Appropriate band-pass filters were selected for each channel: blue channel—D480/60 M (AHF Analysentechnik, Tübingen, Germany); green channel—BG39 glass (3 mm thick; Schott AG, Mainz, Germany); and red channel—HQ 610/75 M-2P (AHF Analysentechnik). High-resolution overview image stacks were acquired using a ×20 objective (XLUMPlanFI ×20/0.95, Olympus) at 850 nm excitation wavelength (Fig. 1, configuration A1). Calcium imaging was performed using a ×40 objective (LUMPlanFI/IR ×40/0.80W, Olympus). When measuring calcium transients in GFP-positive neurons, the signal-to-noise ratio in the functional (i.e., green) channel could be improved by using 820 nm excitation wavelength, which minimized GFP fluorescence contribution in the green channel (Fig. 1, configuration A2).
Fluorescence excitation and detection beam paths used for two-photon imaging with different fluorophore combinations. Dyes in bold: visible; non-bold: invisible with the respective excitation/detection scheme. Abbreviations: λ ex excitation wavelength, PMT photo-multiplier tube, BP band-pass filter, DC dichroic mirror, SP short-pass, LP long-pass. Filter wavelengths are specified in nanometers. For BP filters, the center wavelength and the width of the transmission range are given
Tissue preparation
After in vivo experiments, mice were deeply anesthetized by injection of ketamine (0.1 mL, 50 mg/mL, i.p.). Following injection of 0.05 mL heparin into the left ventricle, animals were intracardially perfused with 10–20 mL of PBS (0.1 M, pH 7.3, 4°C) at 12 mL/min, followed by 20 mL of paraformaldehyde solution (PFA; 4% in 0.1 M PBS, pH 7.3, 4°C) at 12 mL/min. In experiments with YC3.60-expressing mice, animals were subsequently perfused with 5 mL of warm agarose solution (type III-A, Sigma; 1.5% in 0.1 M PBS). This prevented small capillaries from collapsing, which later provided crucial landmarks for cell matching. The brain was then removed from the skull and postfixed in 4% PFA at 4°C overnight. Then the brain was rinsed three times with PBS (0.1 M, pH 7.3, room temperature) and a tissue block containing the area of craniotomy was excised and stored in PBS (0.1 M, pH 7.3, containing 0.04% sodium azide NaN3 to prevent microbial growth) at 4°C until further processing.
Cutting procedure and slice selection
A large field-of-view surface map (average intensity projection of an image stack with 20 μm z-steps) of the fixed tissue block was acquired under the two-photon microscope using a ×4 objective (UPlanFL N ×4/0.13, Olympus) and 850 nm excitation wavelength (Fig. 1, configuration A1). The tissue block was afterwards kept in 30% sucrose at 4°C until sedimented. It was then embedded in TissueTek O.C.T.TM Compound (Sakura Finetek, Alphen aan den Rijn, The Netherlands) and frozen at −20°C before being cut into 140–200-μm-thick coronal slices using a cryostat (both sample and blade temperature were kept at −16°C to avoid tissue fragmentation during sectioning). Sections were mounted vertically between two cover slips (No. 1, Menzel-Gläser, Braunschweig, Germany) using two strips of cover slip as spacer. The pial brain surface segment of the slice was then imaged under the two-photon microscope (Fig. 1, configuration A1) using the ×4 objective mentioned above. In order to identify the slices covering the volume of interest, images of the pial slice surfaces were overlaid onto the surface map acquired before sectioning using Photoshop CS 3 (Adobe Systems, Mountain View, CA, USA).
Immunohistochemistry
Slices were blocked in carrier solution (10% normal donkey serum (Jackson ImmunoResearch, West Grove, PA, USA), 2% Triton X-100 (Sigma-Aldrich, Buchs, Switzerland), and 0.04% NaN3 in 0.1 M PBS) overnight at room temperature on either a shaker (IKA-VIBRAX-VXR, IKA, Staufen, Germany) or a rotation incubator (CMV-ROM, Fröbel Labortechnik, Lindau, Germany). Slices were then incubated with primary antibodies (Table 1) in carrier solution for 3 days at room temperature on the shaker. Afterwards, they were briefly rinsed three times with 0.1 M PBS, then washed three times with PBS for 30 min, and subsequently incubated with secondary antibodies (Table 1) in carrier solution for 1 day at room temperature on the shaker or rotation incubator, respectively. After further rinsing and washing (three times each), slices were cleared in a sequence of sucrose solutions (15%, 30%, 45% and 60% of sucrose in 0.1 M PBS/2% Triton X-100) with 4-h periods per sucrose concentration. Negative controls were separately performed for each secondary antibody. No false-positive candidate cell bodies were detected.
Brain slice imaging and image stack alignment
After immunostaining, brain slices were mounted on microscopy slides in 60% sucrose solution (see above) and protected with cover slips using strips of cover slips as spacers. For each slice, two image stacks (3 μm z-steps) of the region corresponding to the in vivo reference image stack were acquired using the two-photon microscope with a ×20 objective (see above). At 850 nm excitation wavelength, triple immunostained brain slices from GAD67-GFP mice were imaged with the same filter set as for in vivo imaging (Fig. 1, configuration A3). For excitation at 720 nm, a different band-pass filter was used for the blue channel (EP 450/50, AHF Analysentechnik, Tübingen, Germany; Fig. 1, configuration A4). For slices from YC3.60-expressing mice, we used a different detector configuration (Fig. 1, configuration B) with another dichroic mirror (Q 650 LP from AHF Analysentechnik) and an appropriate filter combination for the infrared channel (BrightLine HC 697/75 in series with a Semrock FF01-750/SP-25 infrared block filter, both from AHF Analysentechnik).
The exact locations for stack acquisition were identified using surface blood vessel cross-sections as landmarks. Image stacks were resliced using ImageJ [1] to obtain a 90° rotation. Further rotational fine-tuning was carried out using the TransformJ plug-in (kindly provided by Erik Meijering, Rotterdam) for ImageJ. In detail, a series of rotations around the horizontal x- and y-axes were performed in a systematic trial-and-error approach until optimal rotation fitting was achieved, including scaling and translation operations if required.
Data analysis
Custom-written LabVIEW software was used for in-vivo-to-fixed-slices cell matching and cell annotation. Two-photon calcium imaging movies acquired from the functional channel were registered using the ImageJ plug-in TurboReg [69]. Calcium signals from cells of interested were extracted using ImageJ and further processed with Excel (Microsoft Corporation, Redmont, WA, USA) and Igor Pro 5 (WaveMetrics Inc., Portland, OR, USA). In detail, the procedure was as follows: First, the mean background value was determined inside a blood vessel and subtracted from the movie frames. Then, a baseline image F 0 was defined as the average of the first 20 frames. Relative percentage fluorescence changes (ΔF/F 0) were calculated pixel-wise by subtracting F 0 from the image time series and dividing the result by F 0. Single-cell regions of interest were drawn by hand such that their contours were within the visually perceived cell boundary (with at least one pixel distance to it, in order to exclude surrounding neuropil).
Results
In vivo discrimination of excitatory neurons, GABAergic interneurons, and astrocytes
To broadly discriminate between excitatory and GABAergic neurons, we first employed in vivo two-photon imaging in GAD67-GFP knock-in mice [68], in which GABAergic interneurons express enhanced green fluorescent protein (here simply referred to as GFP). Astrocytes were counterstained by brief application of SR101 to the cortical surface [59]. In addition, cells were unspecifically loaded with the calcium indicator OGB-1 by multi-cell bolus loading (MCBL) [67]. Using a single excitation wavelength (850 nm), SR101-labeled astrocytes were readily identified in the red detection channel, while both OGB-1 and GFP were visible in the blue and the green channel with our optical settings (Figs. 1 and 2a). Despite this cross-talk, GFP-positive cells could be unambiguously identified in triple-labeled populations based on the ratio of fluorescence in the blue and green channel. In a control experiment, we counted GFP-positive cells before and after MCBL of OGB-1 and found that 100% of GFP-positive cells (63 of 63 cells) were identified. This triple-labeling approach thus enabled discrimination of astrocytes, GABAergic interneurons, and excitatory neurons in vivo (Fig. 2b).
In vivo two-photon calcium imaging with discrimination of three major neocortical cell types. a Separate imaging of GFP, Oregon Green 488 BAPTA-1 (OGB-1), and sulforhodamine 101 (SR101) in three independent experiments, in which only one fluorophore was present (detection scheme A1, Fig. 1). Scale bars = 50 μm. Top row: GFP-positive GABAergic interneurons in a GAD67-GFP mouse were identified by their large contribution to the blue channel (450–500 nm spectral window), but they are also visible in the green channel. Middle row: The OGB-1 signal is strongest in the green channel (500–590 nm) but also partially bleeds through in the blue channel. Bottom row: SR101-stained astrocytes are detected in the green and red channels but are the only cells visible in the red channel (590–650 nm). b Overlay of simultaneously acquired two-photon images in all three spectral channels in an experiment in which OGB-1 and SR101 labeling were applied to the neocortex of a GAD67-GFP mouse. Astrocytes (yellow), excitatory neurons (green), and inhibitory interneurons (blue) can be readily discriminated with this in vivo triple stain. c Spontaneous calcium transients for all three distinguishable cell types for the example cells numbered in b (850 nm excitation wavelength)
In all three cell types, we observed spontaneous calcium transients (Fig. 2c). Astrocytes showed slow fluctuations of the OGB-1 fluorescence traces consistent with previous reports [36, 59, 76]. In identified excitatory neurons, individual and compound calcium transients were evident with sharp onsets, 10–50% ΔF/F 0 amplitude and slow exponential decays over a few hundred milliseconds (Fig. 2c), consistent with action potential-evoked calcium transients [24, 42, 43, 67, 79]. In contrast, spontaneous calcium transients in GABAergic interneurons were more variable. Some cells displayed individual, sharp calcium transients similar to excitatory neurons while others showed rather slow fluctuations of the OGB-1 fluorescence signal (Fig. 2c). These differences in calcium transient shape might reflect differences in electrophysiological and calcium-buffering properties between subtypes of GABAergic interneurons. Discrimination of these subtypes and their functional signals in vivo will be essential for understanding local microcircuit dynamics. We therefore set out to devise a method to further dissect cell types within neuronal populations characterized by in vivo two-photon calcium imaging.
Identifying coronal brain slices containing a particular cortex region
Our goal was to establish subtype discrimination within the GABAergic population by IHC against neurochemical markers. For this purpose, the brain has to be perfusion-fixed and cut into slices. In principle, coronal, parasagittal, transverse, or surface-tangential slices may be used (see “Discussion”). Here, we decided to section coronally (in our context, the term coronal is used according to the homologous sections of the human brain, i.e., perpendicular to the rostrocaudal axis).
We first aimed at identifying those coronal brain slices that contained a particular cortical volume previously imaged in vivo (Fig. 3). For this purpose, we used GAD67-GFP mice without extra labeling. In addition to an overview camera image of the brain surface vasculature, a high-resolution two-photon reference image stack was acquired in vivo, which defined the volume of interest (Fig. 3a, b). After perfusion fixation of the brain, a large field-of-view surface map of the whole craniotomy area was obtained with the two-photon microscope, with larger blood vessels still clearly visible (Fig. 3c). A tissue block containing the craniotomy area was then cut into coronal slices of 140–200-μm thickness using a cryostat. The resulting slices were individually mounted between two cover slips and positioned vertically under the two-photon microscope to image their segment of pial brain surface (Fig. 3d, e). By matching the surface blood vessel pattern of each slice against the large field-of-view surface map, we were able to identify the slices that contained the volume of interest (Fig. 3d). Only these slices (typically two to three in number) were selected for further processing.
Preparation of fixed coronal brain slices and registration to the volume of interest. a In vivo large field-of-view camera image of the blood vessel pattern at the cortical surface within a craniotomy in a GAD67-GFP mouse. b High-resolution reference stack acquired in vivo with the two-photon microscope from the region indicated by the red box in a, revealing the subpopulation of GFP-expressing cells. c Large field-of-view two-photon image of the cortical surface originally within the craniotomy captured from the whole brain after perfusion fixation. This additional reference map of the surface blood vessels complements the camera reference map acquired in vivo. d Coronal sectioning of the fixed brain. e Coronal slices were mounted between two cover slips with two strips of cover slip glass as spacers (red). The glass assembly was held together by two clips and glued to a metal bearing. The mount was placed vertically under the two-photon microscope to image the pial surface segments. These segments were registered and overlaid to the two-photon blood vessel reference map as shown in d. In this case, slices 11 to 13 were identified as the slices of interest because they cover the volume contained in the in vivo two-photon reference image stack shown in b (red box)
Identifying the same cells and cell populations in brain slices
In the next step, we used GAD67-GFP mice to re-identify in fixed brain slices the same cells that were previously imaged in vivo. Slices selected according to the procedure described above were mounted horizontally and high-resolution two-photon image stacks were acquired in coronal view (XZ view). Fields-of-view corresponded to the in vivo reference image stack (Fig. 4a–c). We employed tissue clearing with a sucrose gradient [72], which allowed us to acquire image stacks throughout the entire slice thickness (up to 200 μm) without significant loss in imaging quality. In contrast to tissue clearing with Murray’s clear [17], sucrose clearing preserved cell-specific localization of GFP fluorescence.
Identifying the same GABAergic neurons in images acquired in vivo and from fixed tissue slices, respectively. a High-resolution reference image stack acquired in vivo from a GAD67-GFP mouse. b Image stacks acquired from coronal slices covering the volume of interest (same slices as in Fig. 3d). c Example coronal XZ view from the marked slice from the stack in b. d Three-dimensional composite view of the in vivo stack and one slice stack following alignment. e Matching the same GFP-expressing neurons in the in vivo reference stack (left) and in the brain slices (right). Note that the brain slice image is an XY view composed from several adjacent slices. Slice borders are indicated by green dashed lines. The dashed red line corresponds to the frame shown in c. Orange boxes highlight example constellations of cells that are easily recognized in both images. Arrow heads point to the same cells as in c. Note that some cells visible on the right are not visible on the left (and vice versa) due to nonlinear tissue distortions that cannot be corrected for by rotation fitting. Focusing through the three-dimensional image stacks is required to obtain the match for each cell. f Fraction of matched GFP-positive cells analyzed in 20-μm bins for the single example volume shown in d. Slice borders are again indicated as dotted green lines. g Pooled analysis of the average fraction of matched cells as a function of distance from the closest cutting plane. The number of slices considered is specified for each bin (slices from four mice; error bars indicate standard error of the mean (SEM))
For slice cutting, the sectioning plane was oriented as perpendicularly as possible to the pial brain surface. Nevertheless, the in vivo reference image stack and the 90°-rotated image stacks acquired from the corresponding coronal slices were usually slightly tilted with respect to each other (Fig. 4d). Therefore, the image stacks from each coronal slice were aligned to the in vivo two-photon reference stack by applying linear transformations (rotating, scaling, and translating). Surface and descending blood vessels served as key landmarks. In spite of slight nonlinear tissue distortions, cells imaged in vivo could readily be identified in the image stacks from brain slices following this alignment procedure. For comparing cellular staining patterns, it was beneficial to view both stacks (obtained in vivo and in fixed slices, respectively) in either XY or XZ view. Local neighborhood relations between cells (e.g., characteristic groups of cells or proximity to prominent blood vasculature features) facilitated identification of the exact same cells in the two data sets (Fig. 4e).
We quantified the fraction of GFP-positive cells that could be re-identified in brain slices in this manner. The overall percentage of manually matched cells depended on the time invested. Matching rates of around 80% could be readily achieved with about 20 h of work (one person) for a cortical volume of about 300–400 μm side length containing approximately a thousand GFP-expressing GABAergic neurons (647, 826, 1,056, and 1,039 cells, respectively; on average about one cell per minute). With more time invested (around 50 h), cells in more difficult regions could also be matched, reaching matching rates between 95% and 100% for such-sized volumes. Essentially, all GFP-positive cells could be identified in the middle of the brain slices, while at the cut slice surfaces, a few cells were either lost or could not be unambiguously identified (Fig. 4f). Even close to the cutting plane, however, high matching rates (>95%) were reached (Fig. 4g). This analysis demonstrates that large cell populations imaged in vivo can be re-identified almost completely in coronal slices following brain fixation.
Subtype discrimination of GABAergic neurons by triple immunostaining
Following successful identification of individual GABAergic neurons in fixed brain slices, we next aimed at discriminating subtypes by IHC. First, we decided on a suitable set of antibodies. In general, antibody combinations can be flexibly chosen as long as antibodies do not cross-react and permit independent signal acquisition, and the resulting staining sufficiently penetrates the tissue. Here, we chose three classical interneuron markers that are commonly used to define multiple subtypes, namely, the calcium-binding proteins parvalbumin (PV), calretinin (CR), and calbindin (CB) [20, 41, 54]. Primary antibodies were taken from different host species, and appropriate fluorescence labels for secondary antibodies were selected to enable multi-color IHC (Table 1 and Online Resource 1a). A critical issue is how well antibodies penetrate into slices because diffusion of high molecular antibodies is hindered by the extracellular matrix. To test antibody penetration, we analyzed the depth dependence of antibody stain and found an exponential decline of intensity with distance from the slice border (space constant 25 μm for anti-PV stain; Online Resource 1b). We chose 140-μm-thick slices, which enabled sufficient staining of cell bodies in the slice center (Online Resource 1c–d) while providing enough blood vessel surface pattern for convenient slice selection.
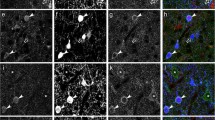
Following antibody staining, we acquired stacks of two-photon images from horizontally mounted coronal slices using three-channel fluorescence detection. The red anti-PV stain marked a subset of GFP-positive cells, all of which were clearly visualized using 850-nm excitation wavelength (Fig. 5a, left). Switching the excitation wavelength to 720 nm, the green anti-CR and blue anti-CB stain could be well discriminated because negligible GFP fluorescence was excited (Fig. 5a, right). With this approach, we were able to distinguish cells that were negative for all three markers and those that contained one or several markers. Figure 5b shows example cells for five different marker combinations: (1) a PV-positive interneuron with multi-polar dendritic morphology, which presumably is a basket cell; (2) a CR-positive cell with typical bipolar dendritic morphology [54]; (3) a CB-positive cell; (4) a neuron positive for both PV and CB; and (5) a triple-negative cell.
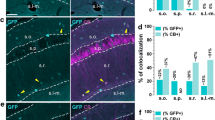
Post hoc triple immunostaining of calcium-binding marker proteins in populations of GABAergic interneurons imaged in vivo. a Superficial cortical layers in a coronal slice stained for parvalbumin (PV), calretinin (CR), and calbindin (CB). Left: At 850-nm excitation, GFP-positive cells are clearly visible as blue cells while PV-positive neurons appear orange. Right: At 720-nm excitation, the three antibody stains against PV, CR, and CB are visible in the red, green, and blue channel, respectively. b Examples of matched GABAergic interneurons with five different combinations of marker proteins expressed (coronal views from fixed slices; upper left insets: in vivo images of the same, GFP-expressing cells). Scale bars = 20 μm. c Three-dimensional view of an image stack of a large population of GABAergic neurons (874 cells) acquired in vivo with color-coded overlay of the classification determined by post hoc immunostaining: PV+ (red), CR+ (green), CB+ (blue), PV+/CB+ (pink), GFP+ only (gray), and GFP+ but not matched (yellow). The estimated extent of the C2 barrel column located by intrinsic optical imaging is indicated with dashed lines. d Layer dependence of the abundance of GABAergic interneurons expressing different protein marker combinations. The percentage fraction of a specific marker combination from all GFP-positive neurons is shown in 50-μm bins for two volumes from two different animals (left: centered to C2 barrel; right: centered to D2 barrel). CR+/− indicates that the presence or absence of CR cannot be specified with certainty due to channel bleed-through under our conditions
The sequence of in vivo imaging, slice cutting, slice selection, immunostaining, and matching can in principle be applied to large volumes, limited mainly by the in vivo penetration depth of the two-photon microscope. In our experiments, we were able to cover volumes as large as the supragranular part of a whole barrel column, covering layer 1 through layer 2/3 with nearly thousand GABAergic interneurons (874 and 842, respectively; Fig. 5c). For two such data sets, we analyzed the depth-dependent distribution of different interneuron subtypes (Fig. 5d). Layer 1 contained nearly exclusively GABAergic cells negative for all three markers. PV-positive cells were more abundant with increasing depth, approaching around 45% of GFP-positive cells at 250 μm depth. CR-positive cells were sparsely found in layer 1 and constituted about 10–20% throughout layer 2. CB-positive cells started to appear at 50–100 μm depths and accounted for 7–14% of all GABAergic cells down to 250 μm, roughly half of this fraction in co-localization with PV. Overall, this depth-dependent marker distribution in superficial cortical layers is in accordance with previous studies [28, 29, 68].
Combining in vivo calcium imaging with post hoc immunostaining
We next applied post hoc IHC to interneurons for which spontaneous calcium transients had been previously measured in vivo similar to Fig. 2. The additional in vivo stains of SR101 (red) and OGB-1 (green) did not interfere with post hoc antibody staining. SR101 was washed out in fixed slices while OGB-1 remained as a diffuse background of much lower intensity than the green anti-CR stain (Fig. 6b). Figure 6a shows example calcium traces measured in vivo from four identified GABAergic neurons as well as four excitatory neurons. The same set of GABAergic cells was identified in the fixed slices. Triple immunostaining revealed that one of the cells was PV-positive, another one CR-positive, and two were triple-negative (Fig. 6b). With this extra information, the fluorescence traces in Fig. 6a could be classified accordingly. Figure 6c displays additional example calcium transients from other identified cells showing these three marker expression patterns. As in Fig. 2c, calcium transients varied in their temporal characteristics. PV-positive interneurons typically showed relatively slow and low-amplitude fluctuations of the calcium signal. CR-positive cells displayed fewer and sharper calcium transients while triple-negative interneurons tended to display larger amplitude and superimposed calcium transients. Attempts to reveal significant differences among the spontaneous calcium transients (e.g., with respect to frequency, amplitude, or shape of transients) that would correspond to the post hoc classification remained, however, inconclusive. Nonetheless, our experiments demonstrate that it is possible to relate functional signals measured in local populations under in vivo conditions to the neuronal subtypes as identified by post hoc IHC.
Post hoc classification of functional in vivo calcium traces from GABAergic interneurons according to their expression profile of calcium-binding proteins. a Left: Field-of-view in barrel cortex of a GAD67-GFP mouse (150 μm depth below the pia), in which two-photon calcium imaging was performed in vivo. Right: Spontaneous calcium transients recorded in the example cells marked with arrows (1–4: GABAergic neurons; 5–8: excitatory neurons). Trace colors indicate the post hoc GABAergic cell classification as revealed in b: light blue, PV−CR−CB−; red, PV+CR−CB−; green, PV−CR+CB−; black, non-GABAergic neurons. b The same area reconstructed from triple immunostained coronal slices by means of rotation fitting with 850-nm (left) and 720-nm (right) excitation. A slice border is indicated as a green dotted line. GFP-positive cells without subtype marker staining appear blue in the left image and are invisible on the right. PV-positive cells are orange in the left and red in the right image. CR-positive and CB-positive cells are green or blue, respectively, in the right image. Matching of GABAergic neurons 1 through 4 with the in vivo image in a is indicated. c Collected examples of spontaneous calcium transients observed in two animals for identified GABAergic neurons with different types of marker combinations. All traces are recorded at different time points except for those marked by arrows and stars, respectively
Post hoc immunostaining of cortical neurons expressing a genetically encoded calcium indicator
Recently, improved genetically encoded calcium indicators have emerged as powerful tools to enable repeated, chronic functional measurements from local neuronal populations [3, 49, 53]. We therefore examined whether our approach can also be applied to cortical populations expressing the genetically encoded calcium indicator Yellow Cameleon 3.60 (YC3.60; [58]). We processed the brains of three wild-type mice, in which neuron-specific YC3.60 expression had been induced by delivery of an adeno-associated viral construct [49] (Fig. 7a). Cell identification in fixed brain slices was possible as in the GAD67-GFP mice, although matching was time-consuming and had to mainly rely on prominent blood vessel curvatures and bifurcations in the densely YC-3.60-labeled populations (Fig. 7a, b). For one experiment, we quantified the matching rate for YC3.60-expressing neurons (>1,000 cells) within three adjacent slices and obtained similarly high values (>99%) as for the GAD67-GFP mouse (Fig. 7c).
Post hoc immunostaining of cortical tissue expressing Yellow Cameleon 3.60 (YC3.60). a In vivo two-photon image of a layer 2/3 neuronal population expressing YC3.60 following viral injection. A particular cell constellation is highlighted by an orange box. b The same cell population in two adjacent fixed slices, rotation-fitted to the in vivo XY view. The anti-PV immunostain is overlaid in red. The green dashed line indicates the slice border. c Fraction of matched cells analyzed for 20-μm bins in a 220 × 370 × 250-μm3 volume (1,427 cells in total). Slice borders are indicated as dotted green lines (slice border B is visible in b). d Coronal side view at the level of the dashed red line in b. The PV-positive cell marked by the arrow head is the same cell marked in the cell constellation in a and b. e Example of anti-PV/anti-CR double immunostaining of YC3.60-expressing cells in the neocortex. In the coronal side view, the bipolar morphology of CR-positive interneurons is evident
For immunostaining, we chose a slightly different set of antibodies that was better compatible with the YC3.60 fluorescence spectrum (Table 1). As an example, anti-PV staining of the cell population shown in Fig. 7a, b revealed one PV-positive interneuron that turned out to be a multi-polar cell at the border of layers 1 and 2 (Fig. 7c). In another experiment, we successfully applied double-labeling IHC to YC3.60-expressing cell populations, identifying both PV-positive and CR-positive subsets of interneurons (Fig. 7d). We conclude that interneuron subtype discrimination with post hoc IHC is possible in local cortical circuits densely labeled with genetically encoded functional markers.
Discussion
In this study, we combined in vivo two-photon calcium imaging with post hoc triple immunostaining to discriminate functional signals in GABAergic neuronal subtypes. We re-identified cells in fixed brain slices by matching corresponding neuronal populations in relatively large volumes. Antibody staining against calcium-binding proteins permitted us to start comparing calcium transients in specific GABAergic subtypes. The method can be combined with both synthetic and genetically encoded calcium indicators. It is therefore particularly well-suited for cell discrimination following chronic in vivo imaging. Post hoc IHC opens novel and flexible possibilities to decompose in vivo network activity patterns into circuit subcomponents and thereby advance our understanding of signal flow through neuronal microcircuits.
Genetic markers versus post hoc immunostaining
Two main approaches currently exist to distinguish different cell types in two-photon calcium imaging studies according to differential gene expression. First, advanced transgenic methods enable expression of fluorescent proteins in specific cell types in vivo (reviewed in [6, 57]). For example, transgenic rodent lines have been produced that express a fluorescent protein either in nearly all GABAergic neurons [68, 73] or in a subset thereof [12, 50, 56, 61]. Moreover, driver lines with cell type-specific expression of the site-specific recombinase Cre can be used to express an arbitrary fluorescent protein either through cross-breeding with a reporter line [52] or through local injection of customized viral shuttles [46]. Genetic fate mapping by conditional activation of Cre recombinase expression at different time points during development is another promising approach to label specific interneuron subtypes [9]. While two-photon calcium imaging in the GAD67-GFP mouse has already been performed in several studies ([42, 65, 77]; our study), the combination with Cre lines driving fluorophore expression in subclasses of GABAergic neurons is just emerging [63, 80]. A major advantage of transgenic approaches is that specific cells can be identified and targeted during the in vivo imaging experiment. In particular, imaging of local population activity may be combined with targeted electrophysiological recordings [16, 26]. On the other hand, production of transgenic mouse lines is still time-intensive [52] and the possibilities to combine multiple fluorescent labels and indicators are limited.
The second, IHC-based approach detects cell-specific marker proteins post hoc in fixed tissue, as shown here and in a few recent studies for GABAergic neurons [42, 60, 74]. This method is not limited to GABAergic neurons or the three types of markers used in our study. First, it can be applied to any mouse line expressing a fluorescent protein in a cell type-specific manner in order to further dissect the labeled cell population. Second, as we have demonstrated by applying the method to YC3.60-expressing cortical tissue, it also masters situations where no sparse candidate cells are present, but rather all cells are more or less equally fluorescent. The method could thus even be applied to other species, for which transgenic lines with cell type-specific labeling are not available [14, 34, 39]. Third, the set of primary antibodies used can in principle be freely chosen, as long as penetration depth and sensitivity is compatible with the chosen thickness of sections. The key advantages of post hoc IHC are its high flexibility, combinatorial application of multiple fluorophores, and that both the target antigen and the fluorescence labels can be exchanged quickly. A disadvantage of the IHC-based approach is, however, that staining is available only post mortem. Unless other readily accessible features such as typical cell morphology can be correlated to the presence of a marker protein or peptide, the respective cells of interest cannot be identified and targeted in vivo. On the other hand, since the two approaches (genetic vs. immunostaining) are complementary, they could also be combined to first achieve in vivo targeting of one cell type, which then could be followed by a more complete post hoc IHC discrimination of subtypes.
Technical aspects
Fixed tissue can be sectioned in different orientations. While we chose coronal sectioning, tangential sectioning is a reasonable alternative [42]. The two sectioning orientations have different advantages when compared to each other. First, with our approach, a high-resolution side view of the cell morphology is readily available. Especially for cells with dendritic and axonal arborizations extending over several cortical layers, morphology reconstruction is easier in coronal sections as it does not have to follow neural processes over several tangential sections. In addition, coronal sectioning is also compatible with the recent achievement of performing in vitro electrophysiology on acute slices obtained from cortex on which calcium imaging has been previously performed in vivo [37, 44]. For acute slices, coronal or parasagittal sectioning is mandatory because tangential sectioning would largely destroy layer-spanning neurons. Second, with coronal sectioning, the thickness of all slices can be precisely controlled as opposed to tangential sectioning, where the thickness of the first slice is usually not well defined. On the other hand, tangential sections can be cut arbitrarily thin, while the coronal sectioning approach requires a certain slice thickness to enable slice identification based on the pial surface blood vessel patterns. For antigens of low abundance, sections as thin as possible are usually required. In addition, with tangential sectioning, it is easier to ensure that the orientation of slicing closely matches the in vivo imaging plane [42]. We conclude that the two approaches complement each other, so that in future studies one can choose whichever one better fits the experimental requirements.
Optimal rotation fitting of the fixed tissue image stack to the in vivo reference stack greatly simplifies matching cells. This is especially the case for YC3.60-infected cortical volumes, where characteristic 3D constellations of sparse landmarks are missing. While we obtained good results with empirically determining angles for rotation by a trial-and-error approach, automated rotation fitting could be used in the future. Based on a set of at least three matched landmarks, rotation angles can either be determined by iterative optimization algorithms [7] or directly calculated [4, 38, 44]. In addition, matching could be assisted by algorithms suggesting the expected position of a cell in the fixed tissue image stack based on its position in the in vivo reference image stack and the coordinate system transformation defined by the already matched landmarks. Such algorithms could even be extended to suggest the most likely match of available candidate cells based on local energy minimization. Furthermore, image segmentation algorithms could be employed for automated detection of soma positions (e.g., [48]).
Despite the high matching rates we achieved, we observed a slight decrease towards slice borders (Fig. 4e). Two reasons may account for this observation. First, cells cut in halves may be more difficult to identify. Second and more importantly, the local neighborhood, which helps to identify a particular cell, is only partly available at slice borders. Both these underlying problems could be circumvented by rotation fitting the in vivo reference image stack to the image stack acquired from the stained slices and comparing both in coronal XZ view. Especially in combination with a matching assistance algorithm as suggested above, this could further improve the matching rate at slice borders.
With our choice of fluorophores for tagging the secondary antibodies and our detection system, we still experienced some channel bleed-through. The green channel was substantially contaminated by the blue fluorophore (AMCA) and slightly also by the red fluorophore (DyLigth594). Both the choice of fluorophores and filters in the detector system could be further optimized to minimize this bleed-through. For example, the emission spectrum of DyLight405 is slightly blue-shifted compared to AMCA and would thus less contaminate the green channel. Further, the band-pass filter we used in the green channel (BG 39) has a rather broad spectral window. A filter with a narrower window (e.g., a 535/50 band-pass filter) would lead to a purer signal. Alternatively, modern commercially available confocal microscopes provide sophisticated fluorescence excitation and detection schemes with which the contributions of the individual fluorophores can be separated much better than with our custom-built two-photon microscope. We therefore expect that simultaneous immunostaining and discrimination of four instead of three markers without significant channel bleed-through should be possible.
Calcium imaging of GABAergic subtypes
Besides the expression pattern of marker proteins and neuropeptides as revealed by IHC or single-cell RT-PCR [71], the electrophysiological properties of GABAergic interneurons, specifically their action potential firing characteristics, are key criteria for classification. Despite controversy in the field, some correlations between the presence of certain markers and the firing pattern of a GABAergic neuron have been found [10, 11, 32, 54]. Because calcium concentration changes evoked by action potentials depend on action potential shape, abundance of voltage-dependent calcium channels, as well as the presence of calcium-binding proteins, action potential-evoked calcium transients in the soma presumably differ between subtypes of GABAergic neurons [15, 21]. In particular, PV is expressed in fast-spiking GABAergic neurons, which exhibit short action potentials and can fire at high frequencies [40, 55]. Compared to cortical pyramidal neurons, PV-positive neurons elicit elementary somatic calcium transients with an about tenfold smaller amplitude [23, 42]. One might therefore expect that in PV-positive neurons single action potential-evoked calcium transients will be hard to detect. Rather, fluctuations in action potential frequency should be reflected in modulation of the fluorescence signal. Our results are at least qualitatively consistent with this expectation. However, similar calcium transients might result from the summation of a few large-amplitude or many small-amplitude calcium transients, respectively. Our failure to observe clear correlations between the presence of GABAergic subtype markers and characteristic features of the corresponding calcium signals thus indicates that it may not be straightforward to classify neurons solely according to their calcium transients, emphasizing the value of post hoc IHC.
Further steps are necessary to understand the relationship between observed calcium transients and the underlying firing patterns throughout an entire local population with all different neuronal types identified. First, a better characterization of elementary somatic calcium transients, evoked by single action potentials, is necessary for the various subtypes of GABAergic interneurons. These data would be best obtained from in vivo measurements and take into account action potential characteristics and calcium-buffering properties. Second, advances in high speed in vivo calcium imaging [13, 31] may help to reveal small but characteristic differences in calcium transient shapes. Third, other neurochemical marker combinations might be better suitable for coarse subtype discrimination. For example, PV, somatostatin (SOM), and vasointestinal peptide have recently been shown to label three non-overlapping subpopulations of the GABAergic cells [42, 78]. Moreover, the combination of PV, SOM, and the ionotropic serotonin receptor 5HT3a has been identified to label 100% of the neocortical GABAergic neurons, distinguishing three major cell groups of different developmental origin [62]. The goal of characterizing the cell type-specific relationships between calcium indicator fluorescence changes and underlying action potentials would be to enable reconstruction of neural spike trains from fluorescence recordings also in the diverse GABAergic interneuron population. Only this will provide a full picture of the temporal sequence of suprathreshold activation of the various components of a local cortical circuit under in vivo conditions.
Future prospects
In recent years, in vivo two-photon calcium imaging of neocortical populations has advanced significantly along several lines. First, new scanning methods have been developed that permit measurements from larger populations in three dimensions and/or at higher effective cell sampling rate [13, 27, 31]. Application of post hoc IHC methods to populations monitored with these specialized techniques should help to obtain a comprehensive picture not only of the distribution of neuronal tuning properties within local networks but also of network dynamics occurring on a short timescale. Second, two-photon imaging is now increasingly applied to behaving animals under either head-restrained or freely behaving conditions [3, 18, 19, 60, 64]. This type of experiment opens entirely new directions for examining the neural correlates of certain behaviors or—vice versa—the behavioral relevance of activity in particular cell types. Post hoc IHC can be expected to have a large impact for this research field because it allows assigning behavior-specific activity patterns to particular GABAergic subtypes and thus should help expand on recent findings of differential interneuron activation in an electrophysiological study [26]. Finally, genetically encoded calcium indicators now enable chronic repeated imaging of the same cells over extended periods of time, sometimes over months [3, 53, 70]. At the end of such longitudinal experiments, post hoc IHC would offer the unique opportunity to gain additional information about particularly interesting sub-ensembles of neurons—identified during the course of an experiment—especially if these constitute only a small fraction of the population. In conclusion, we expect post hoc IHC to become a valuable method for assigning specific cell types to functional neuronal signals and thereby contributing to new insights into the fine-scale functional organization of cortical microcircuits.
References
Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophot Int 11(7):36–42
Alitto HJ, Dan Y (2010) Function of inhibition in visual cortical processing. Curr Opin Neurobiol 20(3):340–346
Andermann ML, Kerlin AM, Reid RC (2010) Chronic cellular imaging of mouse visual cortex during operant behavior and passive viewing. Front Cell Neurosci 4:3
Arun KS, Huang TS, Blostein SD (1987) Least-squares fitting of two 3-D point sets. IEEE Trans Pattern Anal Mach Intell 9(5):698–700
Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsáki G, Cauli B, Defelipe J, Fairén A, Feldmeyer D, Fishell G, Fregnac Y, Freund TF, Gardner D, Gardner EP, Goldberg JH, Helmstaedter M, Hestrin S, Karube F, Kisvárday ZF, Lambolez B, Lewis DA, Marin O, Markram H, Muñoz A, Packer A, Petersen CC, Rockland KS, Rossier J, Rudy B, Somogyi P, Staiger JF, Tamas G, Thomson AM, Toledo-Rodriguez M, Wang Y, West DC, Yuste R (2008) Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci 9(7):557–568
Bernard A, Sorensen SA, Lein ES (2009) Shifting the paradigm: new approaches for characterizing and classifying neurons. Curr Opin Neurobiol 19(5):530–536
Burke EK, Kendall G (eds) (2005) Search methodologies: introductory tutorials in optimization and decision support techniques. Springer, New York
Burkhalter A (2008) Many specialists for suppressing cortical excitation. Front Neurosci 2(2):155–167
Butt SJ, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G (2008) The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron 59(5):722–732
Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K, Hestrin S, Rossier J (1997) Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci 17(10):3894–3906
Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E (2000) Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc Natl Acad Sci USA 97(11):6144–6149
Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, Huang ZJ (2004) Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci 24(43):9598–9611
Cheng A, Goncalves JT, Golshani P, Arisaka K, Portera-Cailliau C (2011) Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nat Methods 8(2):139–142
Ch'ng YH, Reid RC (2010) Cellular imaging of visual cortex reveals the spatial and functional organization of spontaneous activity. Front Integr Neurosci 4:20
Cho KH, Jang JH, Jang HJ, Kim MJ, Yoon SH, Fukuda T, Tennigkeit F, Singer W, Rhie DJ (2010) Subtype-specific dendritic Ca2+ dynamics of inhibitory interneurons in the rat visual cortex. J Neurophysiol 104(2):840–853
Crochet S, Poulet JF, Kremer Y, Petersen CC (2011) Synaptic mechanisms underlying sparse coding of active touch. Neuron 69(6):1160–1175
Dodt HU, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgansberger W, Becker K (2007) Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4(4):331–336
Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW (2007) Imaging large scale neural activity with cellular resolution in awake mobile mice. Neuron 56(1):43–58
Dombeck DA, Graziano MS, Tank DW (2009) Functional clustering of neurons in motor cortex determined by cellular resolution imaging in awake behaving mice. J Neurosci 29(44):13751–13760
Druga R (2009) Neocortical inhibitory system. Folia Biol (Praha) 55(6):201–217
Evstratova A, Chamberland S, Topolnik L (2011) Cell type-specific and activity-dependent dynamics of action potential-evoked Ca2+ signals in dendrites of hippocampal inhibitory interneurons. J Physiol (London) 589:1957–1977
Fishell G, Rudy B (2011) Mechanisms of Inhibition within the telencephalon: "where the wild things are". Annu Rev Neurosci 34:535–567
Franconville R, Revet G, Astorga G, Schwaller B, Llano I (2011) Somatic calcium level reports integrated spiking activity of cerebellar interneurons in vitro and in vivo. J Neurophysiol 106(4):1793–1805. doi:10.1152/jn.00133.2011
Garaschuk O, Milos RI, Grienberger C, Marandi N, Adelsberger H, Konnerth A (2006) Optical monitoring of brain function in vivo: from neurons to networks. Pflugers Arch - Eur J Physiol 453(3):385–396
Garaschuk O, Milos RI, Konnerth A (2006) Targeted bulk-loading of fluorescent indicators for two-photon brain imaging in vivo. Nat Protoc 1(1):380–386
Gentet LJ, Avermann M, Matyas F, Staiger JF, Petersen CC (2010) Membrane potential dynamics of GABAergic neurons in the barrel cortex of behaving mice. Neuron 65(3):422–435
Göbel W, Kampa BM, Helmchen F (2007) Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Methods 4(1):73–79
Gonchar Y, Burkhalter A (1997) Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex 7(4):347–358
Gonchar Y, Wang Q, Burkhalter A (2007) Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat 1:3
Grewe BF, Helmchen F (2009) Optical probing of neuronal ensemble activity. Curr Opin Neurobiol 19(5):520–529
Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F (2010) High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat Methods 7(5):399–405
Gutch H, Battaglia D, Karagiannis A, Gallopin T, Cauli B (2010) Fuzzy classification and inference of interneuronal types. FENS Abstr 5:027.23
Haider B, McCormick DA (2009) Rapid neocortical dynamics: cellular and network mechanisms. Neuron 62(2):171–189
Heider B, Nathanson JL, Isacoff EY, Callaway EM, Siegel RM (2010) Two-photon imaging of calcium in virally transfected striate cortical neurons of behaving monkey. PLoS One 5(11):e13829
Helmchen F, Denk W (2005) Deep tissue two-photon microscopy. Nat Methods 2(12):932–940
Hirase H, Qian L, Bartho P, Buzsaki G (2004) Calcium dynamics of cortical astrocytic networks in vivo. PLoS Biology 2(4):E96
Hofer SB, Ko H, Pichler B, Vogelstein J, Ros H, Zeng H, Lein E, Lesica NA, Mrsic-Flogel TD (2011) Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat Neurosci 14(8):1045–1052
Horn BKP (1987) Closed-form solution of absolute orientation using unit quaternions. J Opt Soc Am 4(4):629–642
Kara P, Boyd JD (2009) A micro-architecture for binocular disparity and ocular dominance in visual cortex. Nature 458(7238):627–631
Kawaguchi Y, Kubota Y (1993) Correlation of physiological subgroupings of nonpyramidal cells with parvalbumin- and calbindinD28k-immunoreactive neurons in layer V of rat frontal cortex. J Neurophysiol 70(1):387–396
Kawaguchi Y, Kubota Y (1997) GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex 7(6):476–486
Kerlin AM, Andermann ML, Berezovskii VK, Reid RC (2010) Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron 67(5):858–871
Kerr JN, Greenberg D, Helmchen F (2005) Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci USA 102(39):14063–14068
Ko H, Hofer SB, Pichler B, Buchanan KA, Sjostrom PJ, Mrsic-Flogel TD (2011) Functional specificity of local synaptic connections in neocortical networks. Nature 473(7345):87–91
Komai S, Denk W, Osten P, Brecht M, Margrie TW (2006) Two-photon targeted patching (TPTP) in vivo. Nat Protoc 1(2):647–652
Kuhlman SJ, Huang ZJ (2008) High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One 3(4):e2005
Langer D, van ’t Hoff M, Helmchen F (2008) HelioScan—a highly versatile control software for laser-scanning microscopes written in LabVIEW. http://www.helioscan.org
Li G, Liu T, Nie J, Guo L, Chen J, Zhu J, Xia W, Mara A, Holley S, Wong ST (2008) Segmentation of touching cell nuclei using gradient flow tracking. J Microsc 231(Pt 1):47–58
Lütcke H, Murayama M, Hahn T, Margolis DJ, Astori S, Zum Alten Borgloh SM, Göbel W, Yang Y, Tang W, Kugler S, Sprengel R, Nagai T, Miyawaki A, Larkum ME, Helmchen F, Hasan MT (2010) Optical recording of neuronal activity with a genetically-encoded calcium indicator in anesthetized and freely moving mice. Front Neural Circuits 4:9
Ma Y, Hu H, Berrebi AS, Mathers PH, Agmon A (2006) Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci 26(19):5069–5082
Ma WP, Liu BH, Li YT, Huang ZJ, Zhang LI, Tao HW (2010) Visual representations by cortical somatostatin inhibitory neurons—selective but with weak and delayed responses. J Neurosci 30(43):14371–14379
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13(1):133–140
Mank M, Santos AF, Direnberger S, Mrsic-Flogel TD, Hofer SB, Stein V, Hendel T, Reiff DF, Levelt C, Borst A, Bonhoeffer T, Hubener M, Griesbeck O (2008) A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods 5(9):805–811
Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C (2004) Interneurons of the neocortical inhibitory system. Nat Rev Neurosci 5(10):793–807
McCormick DA, Connors BW, Lighthall JW, Prince DA (1985) Comparative electrophysiology of pyramidal and sparsely spiny stellate neurons of the neocortex. J Neurophysiol 54(4):782–806
Meyer AH, Katona I, Blatow M, Rozov A, Monyer H (2002) In vivo labeling of parvalbumin-positive interneurons and analysis of electrical coupling in identified neurons. J Neurosci 22(16):7055–7064
Miyoshi G, Fishell G (2006) Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol 16(5):577–584
Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A (2004) Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A 101(29):10554–10559
Nimmerjahn A, Kirchhoff F, Kerr JN, Helmchen F (2004) Sulforhodamine 101 as a specific marker of astroglia in the neocortex in vivo. Nat Methods 1(1):31–37
O'Connor DH, Peron SP, Huber D, Svoboda K (2010) Neural activity in barrel cortex underlying vibrissa-based object localization in mice. Neuron 67(6):1048–1061
Oliva AA Jr, Jiang M, Lam T, Smith KL, Swann JW (2000) Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci 20(9):3354–3368
Rudy B, Fishell G, Lee S, Hjerling-Leffler J (2011) Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol 71(1):45–61
Runyan CA, Schummers J, Van Wart A, Kuhlman SJ, Wilson NR, Huang ZJ, Sur M (2010) Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron 67(5):847–857
Sawinski J, Wallace DJ, Greenberg DS, Grossmann S, Denk W, Kerr JN (2009) Visually evoked activity in cortical cells imaged in freely moving animals. Proc Natl Acad Sci USA 106(46):19557–19562
Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T (2007) GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci 27(8):2145–2149
Soltesz I (2005) Diversity in the neuronal machine—order and variability in interneuronal microcircuits. Oxford University Press, New York
Stosiek C, Garaschuk O, Holthoff K, Konnerth A (2003) In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci USA 100(12):7319–7324
Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T (2003) Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol 467(1):60–79
Thévenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7(1):27–41
Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL (2009) Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6(12):875–881
Toledo-Rodriguez M, Goodman P, Illic M, Wu C, Markram H (2005) Neuropeptide and calcium-binding protein gene expression profiles predict neuronal anatomical type in the juvenile rat. J Physiol 567(Pt 2):401–413
Tsai PS, Friedman B, Ifarraguerri AI, Thompson BD, Lev-Ram V, Schaffer CB, Xiong Q, Tsien RY, Squier JA, Kleinfeld D (2003) All-optical histology using ultrashort laser pulses. Neuron 39(1):27–41
Uematsu M, Hirai Y, Karube F, Ebihara S, Kato M, Abe K, Obata K, Yoshida S, Hirabayashi M, Yanagawa Y, Kawaguchi Y (2008) Quantitative chemical composition of cortical GABAergic neurons revealed in transgenic venus-expressing rats. Cereb Cortex 18(2):315–330
Wallace DJ, Kerr JN (2010) Chasing the cell assembly. Curr Opin Neurobiol 20:296–305
Wang XJ (2010) Neurophysiological and computational principles of cortical rhythms in cognition. Physiol Rev 90(3):1195–1268
Wang X, Lou N, Xu Q, Tian GF, Peng WG, Han X, Kang J, Takano T, Nedergaard M (2006) Astrocytic Ca2+ signaling evoked by sensory stimulation in vivo. Nat Neurosci 9(6):816–823
Wilson JM, Dombeck DA, Diaz-Rios M, Harris-Warrick RM, Brownstone RM (2007) Two-photon calcium imaging of network activity in XFP-expressing neurons in the mouse. J Neurophysiol 97(4):3118–3125
Xu X, Roby KD, Callaway EM (2010) Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol 518(3):389–404
Yaksi E, Friedrich RW (2006) Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nat Methods 3(5):377–383
Zariwala HA, Madisen L, Ahrens KF, Bernard A, Lein ES, Jones AR, Zeng H (2011) Visual tuning properties of genetically identified layer 2/3 neuronal types in the primary visual cortex of cre-transgenic mice. Front Syst Neurosci 4:162
Acknowledgments
We thank S. Broadley, D. Göckeritz-Dujmovic, S. Hintermayer, A. Manalastas, and M. Modest for their contribution to developing and applying the immunostaining procedures; F. Cueni for his contribution to data analysis; S. Giger and H. Kasper for excellent technical assistance; K. Vogt for providing the GAD67-GFP mouse line (originally produced by Y. Yanagawa); M. Hasan and S. Kügler for supplying the YC3.60 virus; D. Margolis and K. Schulz for providing the YC3.60-injected mice; as well as J.-M. Fritschy, D. Margolis, K. Martin, K. Schulz, and M. Thallmair for comments on the manuscript. This work was supported by a PhD fellowship to D.L. from the Roche Research Foundation and grants to F.H. from the Swiss National Science Foundation (SNSF; grant 310030–127091), the German-Swiss Research Group “Barrel Cortex Function” (FOR1341, SNSF 310030–130826), and the Swiss SystemsX.ch initiative (project 2008/2011-Neurochoice).
Ethical standards
All experiments were carried out according to the guidelines of the Institute of Laboratory Animal Science at University of Zurich and were approved by the Cantonal Veterinary Office.
Conflict of interest
The authors declare that there are no conflicts of interest associated with the study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
(PDF 281 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Langer, D., Helmchen, F. Post hoc immunostaining of GABAergic neuronal subtypes following in vivo two-photon calcium imaging in mouse neocortex. Pflugers Arch - Eur J Physiol 463, 339–354 (2012). https://doi.org/10.1007/s00424-011-1048-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-011-1048-9