Abstract
Connexins are four-transmembrane-domain proteins expressed in all vertebrates which form permeable gap junction channels that connect cells. Here, we analysed Connexin-43 (Cx43) transport to the plasma membrane and studied the effects of small GTPases acting along the secretory pathway. We show that both GTP- and GDP-restricted Sar1 prevents exit of Cx43 from the endoplasmic reticulum (ER), but only GTP-restricted Sar1 arrests Cx43 in COP II-coated ER exit sites and accumulates 14-3-3 proteins in the ER fraction. FRET-FLIM data confirm that already in ER exit sites Cx43 exists in oligomeric form, suggesting an in vivo role for 14-3-3 in Cx43 oligomerization. Exit of Cx43 from the ER can be blocked by other factors—such as expression of the β subunit of the COP I coat or p50/dynamitin that acts on the microtubule-based dynein motor complex. GTP-restricted Arf1 blocks Cx43 in the Golgi. Lastly, we show that GTP-restricted Arf6 removes Cx43 gap junction plaques from the cell–cell interface and targets them to degradation. These data provide a molecular explanation of how small GTPases act to regulate Cx43 transport through the secretory pathway, facilitating or abolishing cell–cell communication through gap junctions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Connexins are expressed in complex tissue-specific and/or overlapping patterns in most vertebrate cells, including both neurons and glia in the brain. Two hemichannels from contacting cells are assembled at the cell–cell interface to form a gap junction. Gap junctions provide electrical, energetic and functional connectivity via the passage of K+, Na+, Cl−, and Ca2+ ions, ATP, cAMP, cGMP, IP3, and other molecules <1 kDa (Bozhkova et al. 1974; Chailakhyan 1976; Chailakhyan 1990; Loewenstein 1981; Kumar and Gilula 1996; Goldberg et al. 1999). At least 21 connexin isoforms exist in humans (Willecke et al. 2002; Sohl et al. 2004). Major regulation of connexin-based connectivity is thought to occur via the opening and closure of gap junction channels at the plasma membrane (PM). However, living cells constantly synthesize and degrade connexins and thus gap junction coupling is rapidly regulatable by many factors (including extracellular cues) already in membrane traffic pathways at the different transport steps such as endoplasmic reticulum (ER) exit, traffic to the PM, and internalization/degradation. How are connexin expression, degradation and transport to/from the PM regulated in different tissues and organs? Efforts of many groups are directed to answer these and related questions.
In this study, we used a combination of live cell approaches to analyze transport of Cx43 from the ER to the PM, with emphasis on small GTPases acting in the secretory pathway. Sar1 is the small GTPase of the COPII complex which controls transport of transmembrane and lumenal cargo via ER exit sites (Dascher and Balch 1994; Kuge et al. 1994; Aridor et al. 1995; Bonifacino and Glick 2004). COPII-mediated transport of several transmembrane receptors has been shown to involve 14-3-3 family members (O’Kelly et al. 2002; Yuan et al. 2003; Coblitz et al. 2005). However, the cellular functions of these interactions remain unclear. In proteomics screens from brain fractions we routinely obtained 14-3-3 family members as interactors for the C-terminal tail of Cx43 fused to GST. Different 14-3-3 isoforms were shown to interact with integral transmembrane proteins, such as GABAB receptors (Couve et al. 2001) and potassium channel subunits (Rajan et al. 2002). 14-3-3 machinery has been suggested to regulate the cell surface expression of NMDA and AMPA receptors (Xia et al. 2001). Cx43 harbors at its cytosolic C-terminus serine repeats that are potential 14-3-3 binding sites (i.e. when these sequences are phosphorylated). Interestingly, the serine repeat region at the Cx43 C-terminus is followed by a RPR motif. RxR-type sequences have been shown to act as an ER retention signal in KATP channel subunits (Zerangue et al. 1999). Similar to KATP channel subunits, truncation of this potential retention signal in the Cx43Δ257 mutant leads to increased cell surface appearance, suggesting that the C-terminus is involved in regulating Cx43 transport and/or ER retention.
The fact that ER exit of Cx43 can be blocked by dominant-negative Sar1 was described (Thomas et al. 2005), and also shown by us (Butkevich 2004). But these studies did not discriminate specific actions of the GTP- versus GDP-restricted forms of Sar1 on the state of Cx43. Second, a paper from the Warn-Cramer group demonstrated 14-3-3 interactions with a Cx43 C-terminal motif, using in vitro binding studies and a detailed in silico reconstruction based on the 14-3-3 crystal structure and modeled Cx43 C-terminal peptides (Park et al. 2006). However, a cellular link between all three partners—Sar1, 14-3-3 molecules and Cx43 and the potential role of the RPR sequence in Cx43 transport remained obscure. Cx43 transport from the ER and its oligomerization into hexameric hemichannels has been addressed in work of the Musil group (Musil and Goodenough 1990; VanSlyke et al. 2009), and the Koval group (Maza et al. 2005). Both approaches were based on cross-linking experiments and subcellular fractionation and the data suggested that Cx43 oligomerization may occur during ER exit or shortly thereafter, or in some cases as late as in the trans-Golgi. Here, we combined fluorescence microscopy analyses, FLIM-FRET, cryo-electron microscopy and immunodetection to discriminate effects of different mutants of small GTPases on Cx43 transport and Cx43 oligomerization in living cells.
Materials and methods
Cell culture, molecular biology and transfection of Vero cells
Vero cells (Green monkey kidney fibroblast cells) and primary rat astrocytes were grown at 37°C in DMEM medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin in 10% CO2; 100 μg/ml G418 was added to transfected Vero cells in case of selection for stable transfectants. All cells were transfected by electroporation as described previously (Butkevich et al. 2004; Majoul et al. 2006), and plated on poly-d-lysine-coated 25 mm coverslips (Menzel, Germany). For transfection, cells were washed with 2× PBS and transferred into electroporation media. Fluorescent fusion proteins were constructed using the pEGFP, pEYFP or pECFP vectors (Clontech, Palo Alto, CA, USA). cDNAs encoding isoforms of 14-3-3 (epsilon and beta) were isolated from a mouse brain library (Clontech) by PCR amplification, and subcloned into either pEGFP-N1 or pcDNA3. Carboxy terminally tagged Cx43-FPs were described earlier. Electrophysiological characteristics of FP-tagged Cx43 variants were indistinguishable from the untagged proteins (Butkevich et al. 2004). Expression clones for dominant-negative Arf6 and Sar1 mutants were kind gifts from Dr. J. Donaldson (NIH) and Dr. B. Balch (Scripps), respectively. The Lgp120-GFP clone was a kind gift from P. Luzio (Cambridge, UK).
Anti-Cx43 antibodies raised against the carboxy terminal tail peptide were affinity purified by chromatography using peptide coupled to CNBr-activated Sepharose. The specificity of the Cx43 C-terminal antibody was confirmed with peptide-blocking Western blots and immunoprecipitations. The following commercial antibodies were used: anti-pan 14-3-3 (Chemicon) and an isoform-specific anti-14-3-3 antibody (Santa Cruz); anti-GST (Santa Cruz), mouse anti-HA (Sigma), chicken anti-HA-HRP (Abcam), mouse anti-GFP (Chemicon, Upstate), and rabbit anti-GFP (Abcam).
Electron microscopy of ultra-thin cryo-sections by Tokuyasu
Vero cells grown in cell culture dishes were directly fixed with 2% PFA and 0.1% glutaraldehyde in 0.1 M PBS pH 7.4 for 30 min at room temperature. After removal of the fixation solution, cells were post-fixed with 4% PFA and 0.1% glutaraldehyde for 2 h on ice. Ultrathin cryo-sections were prepared according to Tokuyasu (1978). After washing with PBS, cells were embedded in 10% gelatine, cooled on ice and cut into blocks that were infused with 2.3 M sucrose at 4°C and stored in liquid nitrogen. Ultrathin cryo-sections were cut at −110°C using a diamond knife (Diatome) in an ultracryomicrotome (Leica), collected into a 1:1 mixture of 1.8% methyl-cellulose and 2.3 M sucrose. For immunolabeling, sections were incubated with anti-Cx43 or anti-drebrin antibodies for 1 h followed by a 30 min incubation with 10 or 5 nm colloidal gold-labeled goat-anti-rabbit or goat-anti-mouse Fabs (British BioCell) or protein A-Gold as described by us earlier (Butkevich et al. 2004). Images of sections were acquired with a Philips CM120 electron microscope.
Subcellular fractionation of Vero cells
Subcellular fractionation of Vero cells was performed essentially as described (Majoul et al. 1996). ER, PM and Golgi fractions were sedimented at 120,000×g to obtain samples for SDS-PAGE analysis of proteins. Fractions containing Golgi membranes were recognized using an enzyme assay for UDP-galactosyltransferase, ER fractions were detected with rotenone-insensitive cytochrome C-reductase, and anti-KDEL-sequence antibodies (Majoul et al. 1996; Butkevich et al. 2004). Immunoprecipitates from ER or Golgi fractions were resolved by SDS-PAGE, blotted to nitrocellulose and immunoprobed with commercial anti-Cx43 antibodies, or home-made anti-Cx43 antibodies raised against the C-terminal peptide.
Confocal microscopy and image analysis
Plasmids encoding Cx43-CFP, Cx43-GFP, and Cx43-YFP were introduced into Vero cells by electroporation. These fluorescent chimeras are transported to the PM where they form functional gap junction channels. Earlier, using dual patch clamp and electrophysiology we showed that FP-tagged Cx43 has similar properties as wild-type Cx43 (Butkevich et al. 2004). Routinely we obtain 60–80% of cells expressing fluorescent chimeras using an optimized transfection protocol described earlier (Majoul et al. 2006). To image ECFP, EGFP, or EYFP- tagged Cx43 we used different modes of an Olympus FV1000-based confocal setup. Briefly, internal laser lines at 440, 488, 514, or 543 nm, or an external 532 nm laser were used to excite FPs. In some cases we used a Cascade CCD camera attached to the base port of an IX81 microscope and the illumination derived from a Xenon lamp was reflected using a specially coated dichroic mirror (Z488RDC; Chroma Technology) and passing through a high-numerical aperture objective (63×, NA 1.40, or 100×, NA 1.4, Olympus, Melville, NY). Fluorescence emitted from the tagged proteins passed through narrow Semrock filter sets for ECFP, EGFP and YFP (http://www.laser2000.co.uk/semrock/brightline.htm), except for CFP emission, where a BP 485/17 nm Omega Optics filter was used. Emission light was collected by a cooled-CCD (ORCA ER II; Hamamatsu, Bridgewater, NJ, USA). Cells grown on 25 mm Menzel glass coverslips were placed in a custom-made chamber and imaged at 25–37°C. Several representative cells were recorded for each condition with the same intensity settings and averaged for standard error determination. After transfection with a Cx43-FP encoding plasmid cells were plated at high or low densities (105 or 107 cells, respectively) to study transport of Cx43 in cells with or without cell–cell contacts. Transfected cells were analyzed for transport of CX43-FP at different time points after transfection, from 4 to 12 h, as indicated.
FRET-FLIM measurements and image analysis
FRET-Fluorescence Lifetime Measurements (FLIM) were performed by wide-field frequency-domain FLIM microscopy, using a Lambert Instrument FLIM setup (http://www.lambert-instruments.com) attached to an Olympus IX70 microscope. An Argon-Ion laser, intensity-modulated at 75 MHz, was used as the excitation source and images were recorded with a CCD camera using an image intensifier, which was also modulated at 75 MHz. FLIM sequences were obtained using a 40× and 63×/1.4 numerical aperture (NA) oil objectives. CFP was excited with a 440-nm Argon laser line and fluorescence was detected using CFP-YFP FRET filters (http://www.laser2000.co.uk/semrock/fret.htm). Phases of the emitted fluorescence were calculated from the resulting set of images. Vero cells were imaged also by conventional wide-field fluorescence microscopy, to estimate the relative intensity of the Golgi fluorescence. Two different populations of Cx43 molecules were thus resolved: one that exhibits FRET and another that does not, based on the measured phase shift (phase lifetime or τf).
Images were analyzed using the MetaMorph 7.1 program (Universal Imaging, West Chester, PA, USA) with a FRET calculation program. For final presentation all images were transferred into Adobe Photoshop CS2. The IgorPro mathematical program was used for statistical calculations of data acquired using MetaMorph 7.1.
Results
Cx43 exit from the ER is blocked by both GTP- and GDP-restricted forms of Sar1, but only Sar1-GTP induces accumulation of 14-3-3 proteins in the ER
To analyze the effect of small GTPases on Cx43 transport we standardized transfection and live cell analysis protocols. Fast appearance of newly formed Cx43-FP gap junctions starts to be detectable 2–4 h after transfection. Both fluorescence microscopy and electron microscopy data demonstrate that presence of contacts between cells facilitates the formation of Cx43 positive double-membranes, i.e. gap junctions (Fig. 1). On the other hand, absence of cell–cell contacts prevents this polarized accumulation of connexins at the PM and is accompanied by early accumulation of Cx43-FP in perinuclear lysosomal structures (Fig. 1a–d, arrows).
Analyses of Cx43 assembly and exit from the ER. Vero cells were transfected with Cx43-CFP and analyzed for Cx43 transport at different time intervals after transfection (4–12 h). In contrast to non-contacting cells (a–d), activity-dependent transport of Cx43 to the PM is facilitated by the presence of cell–cell contacts (e). Sar1H79G blocks Cx43 exit from the ER (g, f, h) and prevents formation of gap junction plaques (g). In the absence of cell–cell contacts Cx43 from the Golgi pool is delivered to distinct perinuclear dots (a–d), that can be later colocalized to the lysosomal markers (see further Fig. 6). e Classical Tokuyasu ultrathin cryosections of Vero cells transfected with Cx43-GFP (12 h) were probed with anti-GFP antibodies (Invitrogen), followed by protein A-gold detection (PAG; 10-nm gold). Typical gap junction plaque between two contacting membranes is shown; f Ultrathin cryosections (Tokuyasu) of Vero cells co-transfected with Cx43-GFP and Sar1H79G reveal an ER localization. Cells were fixed 12 h after transfection and probed with anti-GFP antibodies (PAG; 10-nm gold). In contrast to control cells (e), immuno-EM analysis of cells expressing Cx43-GFP and Sar1H79G (f) reveals strong accumulation of Cx43 in the perinuclear ER region. g, h In double-transfected cells Sar1H79G blocks exit of Cx43-GFP from the ER and connexins fail to accumulate at the cell–cell interface (g, arrow); compare to control cells transfected only with Cx43-GFP (e). Scale bar 200 nm for EM and 10 μm for fluorescence microscopy images
One of the requirements for fast cell–cell contact formation is the ability of Cx43 to leave the ER. Here, we blocked ER exit by expression of GTP-restricted Sar1, Sar1H79G, or GDP-restricted Sar1, Sar1T39N, and show that ER exit and appearance of Cx43 at the cell–cell interface is prevented with either mutant (Fig. 1g). Classical Tokuyasu ultrathin cryosections of Vero cells transfected with Cx43-GFP (12 h) and probed with anti-GFP antibodies revealed typical gap junction double-membranes between two contacting cells (Fig. 1e; note absence of ER labeling for Cx43 around the nucleus). Both anti-Cx43 and anti-GFP antibodies enabled detection of transfected Cx43-GFP with protein A-gold. In contrast, cryosections of Vero cells co-transfected with Cx43-GFP and Sar1H79G show strong accumulation of Cx43 in the perinuclear ER region rich in vesicular–tubular extensions (Fig. 1f). Typical double membranes with Cx43 reactivity were not found in cells co-transfected with either Sar1 mutant. These results confirm our live cell data that Cx43-GFP remained in the ER and failed to accumulate at the cell–cell interface. Sar1 acts in the early secretory pathway as part of the COPII coat complex. To visualize the COPII coat we used two COPII subunits, Sec13-GFP and Sec23-YFP, co-transfected with either Sar1H79G or Sar1T39N. Accumulation of Cx43-GFP in COPII-positive ER exit sites was seen only in cells co-transfected with Sar1H79G.
To further analyze morphological differences caused by Sar1 mutants we used biochemical analyses and subcellular fractionation of cells co-transfected with either Sar1H79G and Cx43-FP or Sar1T39N and Cx43-FP. ER and Golgi fractions were separated from the cytosol and PM by fractionation and tested for enzymatic activity: Golgi—UDP-Gal-Transferase, ER—Rotenon-Insensitive Cytochrome C-Reductase are overlayed with decreasing total protein concentration as described earlier (Butkevich et al. 2004). Combined ER fractions 7 and 8 were resolved by SDS-PAGE and immunoprobed with anti-Cx43 and Pan-14-3-3 antibodies. Figure 2i shows the following: lane 1: ER fraction from cells co-transfected with Cx43-FP and Sar1T39N, lane 2: control Golgi fraction, lane 3: ER fraction from cells co-transfected with with Cx43-FP and Sar1H79G, lane 4: cytosol as a positive control for 14-3-3. Under these conditions we detect accumulation of 14-3-3 only in the ER fraction from Vero cells co-transfected with Sar1H79G and Cx43 (lane 3) but not from cells co-transfected with SarT39N and Cx43-FP (lane 1). Time-dependent accumulation of 14-3-3 proteins after co-transfection of Cx43-FP and Sar1H79G in the ER fraction is observed (see Fig. 2m, n). As a further control we used an ER fraction isolated from cells transfected with Sar1H79G alone. Here, in the absence of Cx43-FP, we do not see accumulation of 14-3-3 immunoreactivity (Fig. 2i–1). A likely explanation for this result is that Vero cells express only low levels of endogenous Cx43 (compare to Cx43 transfected cells in which Cx43 expression may be increased up to tenfold, as estimated by immunoblot) and none of the neuronal receptors described to bind 14-3-3.
a–h Two distinct states of Cx43 in the ER in Vero cells expressing Sar1T39N or Sar1H79G. Only Sar1H79G accumulates Cx43 in ER exit sites (ERES) (h). This state correlates with accumulation of 14-3-3 molecules in the ER fraction (see k). a–c Cx43-GFP in cells co-transfected with Sar1T39N remains in the ER membranes, does not segregate into exit sites and does not co-localize with the COPII coat (c, overlay). d Immuno-EM analysis of ultrathin cryosections of Vero cells co-transfected with Cx43-GFP and Sar1T39N probed with anti-GFP antibodies and protein A-10 nm gold revealed that Cx43 immunoreactivity was distributed evenly along an extended ER network and not concentrated in the characteristically rounded ERES. A region of interest (Ins 1) is shown enlarged. e Cx43 transmembrane topology and acid sequence at the cytosolic C-terminus of Cx43 harboring serines in a context suitable for phosphorylation, 14-3-3 protein interaction and RPR/COPI binding motifs. f–g Sar1H79G induces colocalization of Cx43 with the Sec13 subunit of the COPII coat. h Tokuyasu cryo-sections of Vero cells co-transfected with Cx43-GFP and Sar1H79G. Immunogold staining with anti-GFP antibodies show ER cisternae and clearly detectable rounded ERES structures that are decorated with Cx43-GFP. A region of interest showing Cx43 concentrated in ERES is shown (Ins 2). Co-localization between Cx43 and the COPII subunit Sec13-YFP is increased in cells co-expressing Sar1H79G (f–g). i Subcellular fractionation of monolayer-grown Vero cells. ER and Golgi fractions were separated from the cytosol and PM fractions and tested for enzymatic activity: UDP-Gal-Transferase for the Golgi; Rotenon-Insensitive Cytochrome C-Reductase for the ER. Total protein concentrations in the fractions are indicated. j–k Proteins from combined ER fractions 7 and 8 (i) were resolved by SDS-PAGE and immunoblotted with anti-Cx43 and Pan-14-3-3 antibodies. Lanes are as follows: 1 ER fraction of cells transfected with Sar1T39N, 2 control Golgi fraction, 3 ER fraction of cells transfected with Sar1H79G, 4 cytosol. ER fractions (7–8 of the gradient, i) usually show upper oligomeric bands of Cx43 (j) with the Cx43 antibodies; these bands are absent in the cytosol fraction (j, lane 4) (k). Note that 14-3-3 accumulation is observed only in the ER fraction of cells transfected with Sar1H79G. 14-3-3 proteins are abundantly present in cytosol, used as a positive control. l Vero cells transfected with Sar1H79G alone do not show accumulation of 14-3-3 immunoreactivity in the ER (l, 1). Accumulation of 14-3-3 proteins in the ER fraction is seen only in cells co-transfected with Cx43 and Sar1H79G (l, 2). m–n Time-dependent accumulation of 14-3-3 proteins in the ER fraction of Vero cells co-transfected with Cx43 and Sar1H79G (4–10 h). The same membrane was reprobed with anti-tubulin as a loading control
Our interest in 14-3-3 as a potential binding partner for Cx43 was triggered when we noticed a non-canonical 14-3-3 binding motif and consensus serine phosphorylation sites at the very C-terminus of Cx43 (Fig. 2e). Second, we repeatedly recovered 14-3-3 proteins from mice and rat brain extracts with Cx43 C-terminus fused to GST in pull-down assays followed by proteomics analysis. The Cx43 sequence predicts that it can be phosphorylated in its cytosolic C-terminal domain and approximately ten different kinases have indeed been demonstrated to phosphorylate Cx43 (Herve et al. 2007). The crystal structure of 14-3-3 proteins suggests the binding for two phosphorylated molecules (reviewed in Fu et al. 2000). Thus, 14-3-3 proteins may assist lateral dimerization of Cx43 and formation of connexin hexamers already in the ER membrane or upon ER exit. Interestingly, Sar1T39N not only prevented segregation of Cx43 into ER exit sites but also increased Cx43 degradation (Fig. 2j, see lower bands in lane 1). Likely, Cx43 is directed for ER-associated degradation when unable to leave the ER. Note that the same blot was re-probed with anti-14-3-3 antibodies to detect accumulation of 14-3-3 (Fig. 2k). Unfortunately, fluorescence imaging of the distribution of 14-3-3 proteins in single cells could not resolve the Sar1H79G-induced 14-3-3 accumulation on ER membranes, as both the available antibodies against 14-3-3 and 14-3-3-FP after transfection gave a strong cytosolic staining against which background the fine ER membrane structures could not discerned. In contrast, 14-3-3 and Cx43 readily colocalize at non-contacting membranes of double-transfected cells (Fig. 4l–m). Even more surprising was the complete absence of 14-3-3 from the cell–cell interface, where Cx43 is readily detectable (see Fig. 4o–p). The intensity scan in Fig. 4q, r (arrow) indicates Cx43 accumulation at the cell–cell interface. We do not know how long the Cx43/14-3-3 complex may persist as a transport intermediate after ER exit, but hypothesize that during gap junction formation at the PM other binding partners such as submembrane cytoskeleton proteins (e.g. drebrin or ZO1) may displace 14-3-3 from the Cx43 tail.
Oligomerized states of Cx43 in ER exit sites resolved by FRET-FLIM microscopy in living cells
First, to confirm that Sar1H79G blocks Cx43 in ER exit sites (ERES) we co-expressed two subunits of the COPII complex, Sec13 and Sec23, with Cx43. In triple-transfected cells expressing Sar1H79G Cx43 colocalized with both Sec13 (Fig. 2f–g) and Sec23 (Fig. 3b, d). Second, we used Vero cells stably expressing p23-CFP to show that ER exit sites are structures different from dispersed Golgi stacks, as Sar1H79G also prevents transport of Golgi resident proteins from the ER. The transmembrane Golgi protein p23 normally resides in Golgi stacks (Fig. 3a). In cells co-transfected with Cx43-GFP, Sec23-YFP and Sar1H79G Cx43 is found in punctate structures representing ER exit sites and partially colocalize with the dispersed structures labeled by p23-CFP (Fig. 3).
Oligomerization of Cx43 resolved in living cells using FRET-FLIM analysis. ER-to-Golgi transport of oligomerized Cx43 requires COPII and functional dynein motor complex. a p23-CFP stably expressed in Vero cells shows a typical Golgi pattern. Such cells were used for further transfection with Cx43-GFP, Sar1H79G and Sec23-YFP (a–d). The cell in the center (a) expresses all four constructs. b Cx43 is blocked in ER exit sites by Sar1H79G which also blocks p23 transport from the ER and thus prevents its Golgi accumulation (middle cell, a). Cx43 co-localizes with the COPII component Sec23-YFP (d). c Composite overlay of three channels: CFP, GFP and YFP. The fluorescence of the three channels was resolved by linear signal unmixing on the FV1000 confocal microscope. The overlay shows that in Sar1H79G expressing cells Cx43-GFP colocalizes with Sec23-YFP in ERES (resolved by EM in Fig. 2h), while the Golgi protein p23-CFP cannot be detected in the Golgi and is dispersed in the ER membrane. e–h Fluorescence Life Time Microscopy (FLIM) measurements in Vero cells co-expressing the Cx43-CFP/Cx43-YFP FRET pair. e Control cells (expressing no Sar1H79G) co-transfected with the donor/acceptor pair Cx43-CFP and Cx43-YFP show accumulation of both fluorescence signals in the Golgi region. Analysis of the distribution of lifetime (τ, given in nanoseconds) in the Golgi region reveals a strong decrease of τ. Lifetime was diminished from 2.5 to 1.9 ns as a result of FRET induced by close proximity of donor/acceptor proteins, due to oligomerization and segregation of connexins. f Wide-field microscopy image confirming strong accumulation of Cx43-FP in the Golgi region. g, h In cells co-transfected with the Cx43-CFP/Cx43-YFP donor/acceptor pair and Sar1H79G, Cx43-FP fails to arrive to the Golgi and instead appears in perinuclear dots corresponding to ERES (for comparison see EM images of ERES in Fig. 2h). h The Cx43-FP labeled perinuclear structures display a similar decrease of fluorescence lifetime (2.3–1.8 ns) as those seen in the control Golgi region (g), as a result of FRET. i–l ER–to-Golgi traffic of Cx43 utilizes the microtubule-based, minus-end directed dynein motor complex. i Overexpression of p50-CFP, the dynamitin subunit of the dynein/dynactin complex, blocks ER-to-Golgi transport of Cx43-YFP, (j); in such cells Cx43 is unable to enter the cis/medial-Golgi detected with antibodies against KDEL-receptor (k). The overlay (l) shows absence of colocalization for Cx43 with the cis/medial-Golgi marker KDEL-receptor
To better understand the state of Cx43 in dispersed, COPII positive ER exit structures we applied FRET-FLIM analysis of living cells. Generally, it is agreed that Cx43 exists in assembled hexameric form in the Golgi and Cx43 readily accumulates in the Golgi region during time-dependent transport after transfection. Here, we used FRET between Cx43-CFP (donor) and Cx43-YFP (acceptor) to obtain control FLIM lifetime data for the oligomerized form of Cx43. Indeed, although the total fluorescence of Golgi structures viewed in conventional microscope appeared bright (Fig. 3f), the lifetime of the Cx43-CFP donor was shortened [see color coding in red (2.6 ns) and blue (1.8 ns) in Fig. 3e]. As derived theoretically and shown experimentally, FRET can occur only at a molecular distance range of 3–7 nm. R o for the CFP- and YFP-pair is known to be 49–52 Å (Tsien 1998; Miyawaki and Tsien 2000). As revealed by the crystal structures of Cx43 (Unger et al. 1999) and GFP (Yang et al. 1996), this close proximity may occur only if Cx43 is in its hexameric oligomerized state. To measure FRET in living Vero cells we set up lifetime FLIM measurements of the donor, Cx43-CFP, in the presence or absence of the acceptor, Cx43-YFP. Control Vero cells co-expressing this Cx43-CFP/Cx43-YFP FRET pair revealed a strong and significant decrease in the Cx43-CFP lifetime in the Golgi region, where donor lifetime was decreased from 2.5 ± 0.19 to 1.9 ± 0.13 ns (Fig. 3e–f). Cx43-CFP co-expressed with Sar1H79G did not show a Golgi accumulation, as expected. Instead, Cx43 accumulated in ER exit sites, where it co-localized with the COPII subunit Sec23-YFP (Fig. 3d). FLIM measurements obtained from Vero cells co-transfected with Sar1H79G and the Cx43-CFP/Cx43-YFP FRET pair revealed a significant decrease in fluorescence lifetime of Cx43-CFP (tau measured with this setup was reduced from 2.5–2.3s to 1.9–1.7 ns as a result of FRET between CFP- and YFP-molecules in close proximity (Fig. 3). In fact, a very similar decrease of the Cx43-CFP lifetime was observed in the Golgi region of control cells (Fig. 3e) and in ER exit sites when Cx43 transport was blocked by SarH79G (Fig. 3g–h). These data obtained in living cells confirm that Cx43 is present in oligomerized form in both structures: in the Golgi region in control cells and in ER exit sites in cells transfected with Sar1H79G.
Exit of oligomeric Cx43 from the ER requires an intact dynein motor complex
Cx43 transport from the ER to the Golgi involves microtubules and the application of nocodazole has been shown to disrupt Cx43 transport (Giepmans et al. 2001). Microtubule-dependent ER-to-Golgi transport is regulated by the minus-end-directed dynein motor (Burkhardt et al. 1997). Here, we overerexpressed the p50-CFP subunit of the dynactin complex, known to disrupt ER-to-Golgi transport (Valetti et al. 1999). In p50-CFP-transfected cells Cx43-YFP remained in ER exit sites and the ER/Golgi intermediate compartment, ERGIC (Fig. 3i–j). Cx43-YFP co-expressed with p50-CFP failed to colocalize with the KDEL-receptor which resides in the cis- and medial-Golgi (Fig. 3k). These data suggest that although Cx43 exists in its oligomeric form, p50 disrupts the interaction of Cx43 vesicles with microtubular dynein motor components and prevents vesicle transport and/or formation.
Overexpression of β-COP, a subunit of the COPI coat, inhibits Cx43 transport
The COPI coat is localized to Golgi membranes, where it drives the budding of transport vesicles that mediate Golgi-to-ER retrieval and intra-Golgi traffic. The coat minimally consists of the small GTPase Arf1 and the heptameric coatomer complex (α-, β-, β′-, γ-, δ-, ε-, and ζ-COP) and disassembles after vesicle formation in preparation for membrane fusion (reviewed in Duden 2003). Little is known about the role of the cytosolic pool of individual COPI subunits. The Cx43 C-terminus harbors a COPI binding motif (RPR) directly after the 14-3-3 binding motif (Fig. 2e, sequences). In Vero cells overexpressing β-COP tagged with CFP (Fig. 4i) Cx43 remained in the ER (Fig. 4j, k). In contrast, a cell (on the left in Fig. 4j, k) separately transfected with Cx43 alone and then co-plated with cells double-transfected with Cx43-YFP and β-COP-CFP shows nice accumulation of Cx43 at the cell–cell interface (Fig. 4k, arrow). As discussed above, the Cx43 C-terminus contains two almost overlapping protein interaction motifs, a potential 14-3-3 binding site with a serine phosphorylation consensus and a RPR sequence. RxR motifs have been shown using the two-hybrid system to interact with β- and δ-COP (Michelsen et al. 2007). It is conceivable that a balance of 14-3-3 and the monomeric β-COP pool is used by the cell for fine tuning of Cx43 exit to the PM- versus ER-associated degradation. Arginine-based COPI binding motifs were shown to act as a ER retention signal for other multisubunit protein complexes such as GABAB G-protein coupled receptors (Margeta-Mitrovic et al. 2000) and the NMDA receptor splice variant NR1 (Standley et al. 2000; Scott et al. 2001). COPI binding acts to prevent the cell surface appearance of both neuronal receptors. Taken together these data lead to the hypothesis that recognition of arginine-based signals for ER retention of neuronal receptors and Cx43 share a common mechanism in higher eukaryotic cells.
Arf1Q71L prevents formation of gap junction plaques and blocks Cx43 exit from the Golgi. Effects of expression of β-COP and 14-3-3 expression on Cx43 transport to the Golgi. a–b Cx43-CFP co-expressed with wild-type Arf1-YFP in Vero cells. Contacting cells form gap junctions (indicated by arrows). In the absence of cycloheximide Cx43-CFP can be seen in the ER, Golgi and at the PM. c–d In cells expressing Arf1Q71L, Cx43-CFP fails to form gap junction plaques at the PM. e, f Cells co-expressing Cx43-CFP and wild-type Arf1-YFP and treated for 1 h with cycloheximide before imaging display Cx43 only at the PM (arrows Cx43-CFP at the cell–cell interface). g, h Cx43-CFP accumulates in the Golgi in cells co-expressing Arf1Q71L. A cell–cell contact region depleted of Cx43-CFP is indicated by arrow in g. Cells were treated for 1 h with cycloheximide. i, j Expression of β-COP-CFP results in a strong block of ER-to-Golgi transport of Cx43-YFP. k Enlarged overlay of the two channels. An ER distribution for Cx43 is observed in the cell to the right that expresses β-COP-CFP. In contrast, the cell on the left transfected only with Cx43-YFP shows a PM labeling for Cx43 at a cell–cell contact site (arrow). l, m, n 14-3-3 colocalizes with Cx43 at the edge of non-contacting PM regions in co-transfected cells. o–p In cells co-transfected with Cx43-CFP and 14-3-3ε-YFP, 14-3-3 reactivity never accumulates at the contacting cell–cell interface. However, the two proteins do co-localize in the Golgi (arrows). q Absence of 14-3-3ε at the cell–cell interface revealed by fluorescence intensity distribution; r In contrast, in the same cell Cx43 is clearly visible seen at the PM interface with a contacting cell (arrow)
The long-lived COPI/coatomer complex (with a measured half-life of ~28 h once assembled; Lowe and Kreis 1996) is composed of seven stoichiometric subunits. Therefore, the β-COPI-CFP subunit when expressed in cells for 10–12 h is incorporated into coatomer only to a limited degree. Nevertheless, 10–15% of transfected cells revealed a clear Golgi staining for β-COP-CFP, thus allowing for statistical analysis of effects on Cx43 transport. Surprisingly, we find that not only Golgi–PM transport of Cx43-YFP was blocked in cells expressing β-COP-CFP but also ER exit of Cx43 is impaired, demonstrating a novel dominant-negative effect of β-COP overexpression on protein transport in the secretory pathway.
GTP-restricted Arf1 blocks Cx43 in the Golgi and prevents formation of gap junction plaques
We further analyzed processing of Cx43 on the way to the PM. Normally Cx43 accumulates in the Golgi complex, where a master regulatory switch for entry and exit of protein cargo, the small GTPase Arf1, operates. Arf1 is a component of the coat of COPI vesicles that mediate Golgi-to-ER retrieval and intra-Golgi trafficking. Arf1 is a very weak GTPase by itself and thus requires ARF-GTPase activating proteins (ARFGAPs) for its functional cycle, of which three different isoforms ARFGAP1-3 are expressed in mammalian cells that together perform essential functions (see Frigerio et al. 2007). Cx43-CFP when co-expressed with wild-type Arf1-YFP rapidly formed gap junctions in contacting Vero cells (Fig. 4a, b), similar to contacting control cells (data not shown). In the presence of cycloheximide Cx43 was localized mainly at the PM (Fig. 4e, f), as both the ER and Golgi pools were rapidly cleared by transport to the PM. In contrast, in cells co-expressing the GTP-restricted ARF1, Arf1Q71L, Cx43-CFP failed to leave the Golgi and gap junction plaques were absent between contacting cells (Fig. 4c, d, g, h). Our data thus confirm that both the COPI complex and the small GTPase Arf1 are involved in Cx43 transport.
GTP-restricted Arf6 reduces the size of gap junction plaques and facilitates Cx43 degradation via a non-lysosomal pathway
The small GTPase Arf6 is involved in traffic in between the PM and endosomes. However, precisely how the activation of Arf6 regulates vesicular transport remains unknown. We here demonstrate a strong reduction of Cx43-positive cell–cell interfaces in Vero cells upon expression of a GTP-restricted Arf6 mutant, Arf6Q67L (Fig. 5a). Control cells transfected with Cx43-GFP alone displayed a normal cell–cell interface localization for Cx43 (Fig. 5b). Expression of a GDP-restricted Arf6 mutant, Arf6T44N, did not alter this localization of Cx43 in gap junctions compared to control cells (data not shown). Both GFP signals were recorded 10 h after transfection. Statistical evaluations of data obtained from randomly taken pictures of monolayer-grown transfected cells confirmed that in cells co-transfected with Arf6Q67L a strong decrease of the gap junctional cell–cell interface occurs. Cx43-GFP gap junction staining was reduced down to 10% of the level observed in control cells. The relative decrease in gap junction surface area induced by Arf6Q67L and mathematical treatment of the data is shown in Table 1. Inverted black–white images of Vero cells transfected with Cx43-GFP alone as a control revealed accumulation of Cx43-GFP at cell–cell interfaces, in the Golgi region, and in perinuclear lysosomes (Fig. 5c). In contrast, in cells co-expressing Arf6Q67L a gap junction-like cell–cell interface typically remained only in small regions (indicated by arrows in Fig. 5d). Note the complete absence of Cx43 accumulation in perinuclear lysosomes in such cells, especially striking when compared to the lysosomal Cx43 accumulation routinely observed in control cells (compare Fig. 5a, d vs. b, c). At the cell periphery Cx43 is partially colocalized with Paxillin in Arf6Q67L-transfected cells (Fig. 5e–g). The membrane fractions of control cells (left) and Arf6Q67L-transfected cells (right) were separated by 10% SDS-PAGE and probed with anti-Cx43 antibodies. Immunoblot analysis indicated strong degradation of Cx43 in the membrane fraction from Arf6Q67L-transfected cells compared to control cells (Fig. 5h). Using live cell time-lapse microscopy we analyzed the attempt and failure of Cx43-GFP in cells co-expressing Arf6Q67L to form gap junction plaques (Fig. 5i, time interval 7–12 h). As two cells were unable to align their cell–cell interface, the few remaining gap junctions dots were degraded. Our results demonstrate that expression of Arf6Q67L strongly diminishes formation of Cx43 gap junctions. Furthermore, cells expressing Arf6Q67L were unable to maintain pre-existing Cx43-GFP gap junction plaques at the cell–cell interface (data not shown). A possible explanation of the strong effect of Arf6Q67L on gap junction formation/maintenance is that Arf6 facilitates endocytosis, removing newly arrived Cx43-GFP from junctional contacts (Fig. 5i).
Arf6-GTP diminishes formation of Cx43 gap junctions and facilitates Cx43 degradation. a Formation of gap junction plaques is strongly reduced in Vero cells co-expressing Cx43-GFP (green) and Arf6Q67L (not visualized here). The GFP signal was recorded 10 h after transfection. b Control cells expressing Cx43-GFP alone display the usual accumulation of Cx43-GFP in gap junctions at the cell–cell interface at 10 h after transfection. Inverted black–white images (c, d) more clearly reveal the relative decrease in cell surface area occupied by Cx43 gap junctions in cells expressing Arf6Q67L. Note absence of lysosomal accumulation of Cx43 in Arf6Q67L expressing cells (d) compared to control cells (c). For comparison lysosomal accumulation of Cx43 colocalized with lysosomal markers in control cells is shown in Fig. 6a–f. e, f, g In Arf6Q67L expressing cells Cx43-GFP (e) partially colocalizes with Paxillin (f) at the cell periphery (overlay in g). Note again absence of Cx43 accumulation in characteristic lysosomal structures. h Membrane fractions of control Vero cells (left) or Arf6Q67L expressing cells (right) were immunoblotted with anti-Cx43 antibodies. Strong degradation of Cx43 in the membrane fraction from ARF6Q67L expressing cells is evident. The upper (phosphorylated) forms of Cx43 are more resistant to degradation (indicated by arrowhead). i Time-dependent analyses of cells co-expressing Cx43-GFP and Arf6Q67L (time interval 7–12 h) reveal the failure of Cx43 to form gap junction plaques at cell–cell interfaces. Remaining gap junctions are degraded (compare to cells expressing Cx43-GFP alone (b–c)). j Diagram of transfection rate and gap junction formation in control cells expressing Cx43-GFP alone (yellow bars) and cells co-expressing Arf6Q67L and Cx43-GFP (violet bars). Note the strong reduction in total gap junction area in the presence of Arf6Q67L
Cx43 redistributes to lysosomes and undergoes increased degradation in response to cell stress
To address the mechanism of Cx43-GFP disappearance from the PM in Arf6Q67L-expressing cells we applied Chloroquine (CLQ) a known inhibitor of lysosomal degradation. Surprisingly, CLQ did not prevent removal and degradation of Cx43 even at high concentrations (Fig. 6). On the other hand, a mixture of proteasomal inhibitors (epoxomicin, MG132, and Lactacystin, each at 5 μM) did partially restore Cx43-GFP accumulation at the cell–cell interface (data not shown), suggesting that under the conditions described Cx43 undergoes proteasomal degradation. In contrast to the effect of Arf6Q67L on Cx43 degradation shown above, cellular stress induced by exposure of Cx43 transfected cells to high potassium medium (120 mM without Ca2+ for 2 h) did trigger lysosomal degradation. Under this condition we detected extensive colocalization of Cx43-CFP in ring-like structures positive for the lysosomal protein Lgp120-YFP (Fig. 6d–f) compared to control cells (Fig. 6a–c). Image analysis confirmed that under this stress condition significantly less Cx43-FP was present at the cell surface and more present in lysosomes when compared to control cells. When we co-expressed Ubiquitin-CFP and Lgp120-YFP we found that both proteins co-localized in a perinuclear region and in specific rings underneath the PM (Fig. 6g–i). Electron microscopy revealed that these structures represent multivesicular bodies (data not shown). Ubiquitin-YFP was frequently found colocalized with Cx43-CFP in rings (Fig. 6m, n). In contrast, the Golgi protein p23-YFP did not colocalize with Ubiquitin-CFP or Lgp120-CFP in co-expressing cells, confirming that the lysosomal and Golgi pools of Cx43-GFP can be distinguished by light microscopy (Fig. 6j, k, i, and data not shown). Cx43-CFP internalized from the non-contacting surfaces of PM co-localized with Ubiquitin-YFP in ring-like structures resembling annular junctions. Similar structures were visible in cells expressing Cx43-GFP upon exposure to Triton X-100 (Fig. 6p), a treatment known to remove all but detergent-resistant membrane structures. Interestingly, lysosomes were not present underneath freshly formed Cx43 plaques (Fig. 6q, r, s), suggesting that lysosomal degradation involves regulated transport and/or internalization of Cx43 residing in mature gap junctions.
Cx43 transport to the cell surface and degradation in lysosomes in response to mild stress. Vero cells were co-transfected with Cx43-CFP (a) and the lysosomal marker Lgp120-YFP (b); images were taken 10 h after transfection via CFP and YFP channels, respectively. Overlay of a and b reveals only partial colocalization in the perinuclear region (c). d–f Cellular stress induced by exposure of transfected cells to high potassium medium (120 mM K without Ca2+) for 2 h redirects Cx43 from the PM to lysosomes. Extensive colocalization of Cx43-CFP (d) and the lysosomal marker lgp-120-YFP (e) in ring-like, typical lysosomal structures is seen in the overlay (f). g, h, i In Vero cells co-expressing Ubiquitin-CFP (g) and Lgp-120-YFP (h) these proteins colocalize in a perinuclear region and in specific rings underneath the PM (i). By EM these structures have the appearance of multivesicular bodies (data not shown). j, k, l In contrast, the Golgi protein p23-YFP (j) does not co-localize with Ubiquitin-CFP (k) upon co-expression (see overlay in l). m, n, o Cx43-CFP internalized from non-contacting PM areas (m, shown in blue) directly co-localized with Ubiquitin-YFP (n, shown in yellow) in ring-like lysosomal structures (displayed enlarged, as red/green overlay, in o). Remarkably, similar types of structures can be observed after brief treatment of cells harboring Cx43-GFP plaques with Triton X-100 (p), known to destroy all but detergent-resistant structures. q, r, s Lysosomes (Lgp120-CFP, q) are not present underneath freshly formed Cx43 plaques (Cx43-YFP, r), see arrow pointing to a Cx43-YFP-positive cell–cell contact region
Discussion
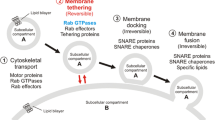
Our data presented here demonstrate that Cx43 traffic from the ER to the PM is strongly regulated by small GTPases. We analyzed how Cx43 can be blocked in the ER and in the Golgi complex, or how it can be removed from the PM, using dominant-negative mutants of the small GTPases Sar1, Arf1, and Arf6 (summarized in cartoon form in Fig. 7). We discovered that the GTP- and GDP-restricted forms of Sar1, Sar1H79G and Sar1T39N, have distinct effects on the cellular fate of Cx43. Using live cell FRET analyses, biochemistry, subcellular fractionation and cryo-immunoelectron microscopy we confirm that only the GTP-restricted form, Sar1H79G, allows segregation of Cx43 from the ER membrane into ER exit sites (ERES). We resolved ERES structures by electron microscopy. A strong accumulation of Cx43 immunoreactivity in ERES was observed in cells expressing Sar1H79G and Cx43 where it co-localized with the COPII subunits Sec13 and Sec23. In contrast, the GDP-restricted mutant Sar1T39N does not allow segregation of Cx43 into ERES. In such cells Cx43 remains smoothly distributed along the ER network. Biochemical analysis revealed that this “non-assembled” state is accompanied by increased degradation of Cx43.
Schematic involvement of small GTPases in Cx43 transport. Left panel Distribution and effects of three small GTPases that act in the biosynthetic pathway, Sar1, Arf1, and Arf6, on the transport of Cx43. Differential blocks induced by GDP- and GTP-restricted Sar1 on ER-to-Golgi transport of Cx43: Sar1T39N prevents assembly into ER exit sites—left, Sar1H79G blocks Cx43 in ER exit sites—right (monomeric and oligomeric forms of Cx43 are depicted in red). Cx43 exit from the Golgi is blocked by ARF1Q71L. Recycling and removal of Cx43 at the PM is Arf6-dependent. This step may be important for regulation of Cx43 at the PM in response to extracellular signals, migration, or initiation of transformation. Middle panel ER-Golgi-PM transport of connexins and formation of gap junctions at the cell–cell interface upon contact. Right panel Possible role of 14-3-3 proteins in Cx43 oligomerization/segregation into ERES
Next, we discovered that GTP-restricted Sar1, Sar1H79G, induces accumulation of 14-3-3 reactivity in the ER fraction in cells co-transfected with Cx43-FP. We analyzed oligomeric states of Cx43 in living cells using FRET between Cx43-CFP (donor) and Cx43-YFP (acceptor) co-expressed in the same cell. Live cell FRET-FLIM analyses confirmed that upon oligomerization of Cx43-FP the donor lifetime was strongly decreased from 2.6–2.4 to 1.9–1.8 ns. The inter-molecular distances between Cx43-FP molecules in ERES calculated from this data are in the range from 3–4 to 6.5 nm (with the Ro for the CFP/YFP-pair known to be 4.9–5.2 nm; Tsien 1998; Burkhardt et al. 1997).
The cytosolic C-terminus of Cx43 contains multiple sequences for phosphorylation and protein–protein interactions which are extensively under study by several groups (Giepmans et al. 2001; Butkevich et al. 2004; Park et al. 2006). We suggest here that close proximity and a correct “oligomerized conformation” of Cx43 induced by Sar1H79G may allow consensus phosphorylation at the Cx43-C-terminus followed by its interactions with 14-3-3 molecules. Indeed, we show that co-expression of Cx43 with Sar1H79G induces accumulation of 14-3-3 proteins in the ER fraction, whereas co-expression of Cx43 with Sar1T39N or Cx43 expression alone does not. The crystal structure of 14-3-3 proteins suggests binding for two phosphorylated molecules (Fu et al. 2000), supporting our view on the possible oligomerization of Cx43 in the ER. Taking into account that 14-3-3 proteins exist in the cytosol as dimers, and that plausible 14-3-3 interactions with a Cx43 C-terminal motif have been reconstructed by the Warn-Cramer group (Park et al. 2006), we speculate that phosphorylation at and 14-3-3 binding to the C-terminus of Cx43 induces its lateral segregation in the ER membrane, oligomerization, and inclusion of these Cx43 assemblies into ERES. These are the molecular events bringing tagged Cx43 molecules into close proximity as seen in our FRET-FLIM data which demonstrate a decreased donor lifetime (Cx43-CFP) in ERES. Significant decrease in the donor lifetime as observed (from 2.5 to 1.9 ns) occurs only when the donor/acceptor-pair, Cx43-CFP/Cx43-YFP, is in close proximity as occurs during oligomerization of Cx43 (see cartoon in Fig. 7, right panel). All connexins, except Cx26, are known to be phosphoproteins. In view of the novel 14-3-3/Cx43 interaction presented here, regulated phosphorylation of the C-terminal domain of Cx43 by protein kinases may be important not only for assembly, closure or opening of channels at the PM, but also for Cx43 exit from the ER. About ten different kinases have been demonstrated to be able to phosphorylate Cx43 (for review see Herve et al. 2007). Thus, a number of different tissue-specific and developmentally regulated kinases are expected to participate in phosphorylation-dependent events.
Uniquely, the C-terminus of Cx43 harbors a 14-3-3 binding motif immediately followed by a potential COPI-dependent retention motif RxR, RPR (Fig. 2e). Here, we tested the biological activity of this motif. We show in living cells that overexpression of the β-subunit of COPI/coatomer prevents ER exit and Golgi transport of Cx43 (Fig. 4i–k). By two-hybrid analysis it has been shown that the RPR sequence is recognized by the β- and δ-COPI subunits and that this interaction is important for protein sorting in yeast (Michelsen et al. 2007). Based on our live cell analyses and in vitro data obtained by others we suggest that oligomerization and segregation of Cx43 in the ER, normally induced by 14-3-3 binding, can be sterically impaired by β-COPI binding. Such a mechanism could be used for fine regulation of Cx43 transport to the PM via signaling. A 14-3-3 binding motif followed by a COPI binding motif was not described for connexins before, but has been shown for some transmembrane receptors and ion channels (Margeta-Mitrovic 2002; Standley et al. 2000; Scott et al. 2001). What happens to Cx43 when it is accumulated in the ER? Locked in the ER, Cx43 may interact with ER ubiquitin-like family proteins as e.g. CIP75 (Li et al. 2008) to be subjected to ER-associated degradation.
Cx43 permeability to small molecules differs in tissues of different origin such as brain, heart, kidney, and skin. We infer a mechanistic role of Cx43/14-3-3/β-COPI interactions observed at the single cell level to explain how the developmental increase in 14-3-3 expression may favor surface appearance of Cx43. Cx43 level is elevated in astrocytes and in early development astrocytes display the highest degree of dye transfer (our unpublished data). 14-3-3 protein expression levels and the type of isoforms expressed vary in specific brain regions and cell types (Baxter et al. 2002), but generally expression is upregulated during post-natal brain development (Toyooka et al. 2002). Similar mechanisms may be triggered during tumorigenesis, where reversely a decrease of 14-3-3 expression may cause ER accumulation of Cx43 and reduced gap junction communication. Our results obtained at the single cell level reveal how 14-3-3 proteins may facilitate Cx43 exit from the ER and suggest that 14-3-3 proteins work in concert with other important molecules of the secretory pathway to bring connexin molecules to the PM.
We show here that even though oligomerization of Cx43 may proceed correctly, the impairment of dynein-dependent transport step out of the ER can cause the same phenotype as impaired oligomerization of Cx43. Expression of the p50/dynamitin subunit of the dynactin complex, as well as microinjection of anti-dynein antibodies (data not shown here), prevented arrival of Cx43 oligomers in the Golgi complex. Strong dependence of the ER-to-Golgi transport step of Cx43 on intact microtubules was observed by us and other groups. Here, we demonstrate that when arrested by p50/dynamitin overexpression, Cx43 resides in dotted ERES-like structures that co-localize with COPII components and resemble those seen in Sar1H79G-transfected cells. In cells expressing p50/dynamitin and fixed for immunofluorescence we were unable to co-localize Cx43 even with the KDEL-receptor, which is cycling between the ER and the cis-/medial-Golgi. We conclude that in p50-transfected cells Cx43 containing structures may remain still associated with the ER membranes, distinct from KDEL-receptor-positive cis-Golgi structures, and, although remaining mobile, fail to accumulate/be loaded on existing intact microtubules (Fig. 4i–l). Since the half-life of Cx43 e.g. in cardiac myocytes is as short as 90 min (Darrow et al. 1996), fast and regulated delivery of Cx43 to the PM may be crucial for cell survival in the heart in vivo. In vitro reconstructions showed that transport of assembled oligomeric proteins along microtubules is generally faster and more progressive than that of their monomeric subunits (Klopfenstein et al. 2002). It remains to be tested whether monomeric connexin subunits can move more effectively along microtubules in living cells.
Normally β-COP and Arf1 co-localize in the Golgi region, as they tightly function together in COPI vesicle budding on Golgi membranes. Upon binding of GTP Arf1 translocates from the cytosol onto Golgi membranes and recruits the coatomer complex (reviewed in Duden 2003). In this study, we demonstrate that Arf1Q71L blocks Cx43 in the Golgi complex, whereas overexpression of wild-type Arf1 allows PM appearance of Cx43. Although we could not resolve 14-3-3 accumulation on the ER membrane against background signal by fluorescence imaging, a clear Golgi pattern of 14-3-3 can be seen during Cx43 accumulation in this organelle. How long does the phosphorylated C-terminus of Cx43 normally remain associated with 14-3-3? Again, cellular analyses of Cx43 distribution with line scans revealed an accumulation of 14-3-3 in the Golgi region (only IF shown here, Fig. 4o–r). Moreover, 14-3-3 proteins can be co-localized with Cx43 at the non-contacting plasma membrane regions, but was never detected at cell–cell interfaces (see Fig. 4). This may be explained by the notion that at the cell–cell interface Cx43 has stronger “stabilizing” interactions (e.g. ZO-1 or drebrin) that displace 14-3-3 from the C-terminus of Cx43.
Lastly, we describe the novel phenomenon of complete and rapid Arf6Q67L-dependent removal of Cx43 gap junctions from the PM. Extracts of cells co-expressing Cx43 and Arf6Q67L were analyzed by immunoblot. Anti-Cx43 antibodies revealed strong degradation of Cx43 induced by Arf6Q67L (Fig. 5). However, the gap junction localization of Cx43-GFP was not affected in cells expressing the GDP-restricted mutant Arf6T44N. What is the biological significance of the Arf6Q67L effect on Cx43 removal at the tissue level? Arf6 action may be required for rapid disassembly of Cx43 plaques in developing or migrating cells as well as during neoplastic transformation (Krutovskikh 2002). This hypothesis may be supported by the observation that at the cell periphery Cx43 is colocalized with paxillin, known to be involved in cell migration processes (Fig. 5e–g). The loss of Cx43-containing gap junctions in Arf6Q67L expressing cells does not involve lysosomal accumulation of Cx43. Only proteasomal inhibitors partially prevented this Arf6Q67L-dependent disappearance of Cx43 from the cell–cell interface. In contrast, lysosomes are readily detectable in control cells in which Cx43-FP alone was expressed. Different GTP- and GDP-restricted Arf6 mutants were extensively studied by the Donaldson group. Arf6Q67L was shown to stimulate the production of PIP2 known to enhance clathrin-dependent endocytosis as well as internalization via clathrin-independent mechanisms (Naslavsky et al. 2004; Jovanovic et al. 2006). A GDP-restricted Arf6, Arf6T27N, was much less potent in its ability to decrease the formation of Cx43 gap junctions than Arf6Q67L (data not shown). A remarkable effect of Arf6Q67L expression was the total absence of lysosomal staining for Cx43-GFP. Although massive degradation should occur after Cx43 internalization, Cx43-positive lysosomal structures were undetectable in cells expressing Arf6Q67L.
Data from different groups on the cellular activities of Arf6 are still controversial. Thus, it was demonstrated that Arf6Q67L expression inhibits transferrin receptor internalization (supposed to occur in clathrin-coated vesicles) without affecting its recycling (Paleotti et al. 2005). Arf6-GTP was shown to interact specifically with the PM-specific adaptor protein complex AP-2 and promotes its membrane recruitment. These findings suggest that Arf6 plays a major role in clathrin-mediated endocytosis by directly controlling the assembly of the AP-2/clathrin coat. Several different functions were assigned to ARF6 including endocytosis, membrane recycling, remodeling of the actin cytoskeleton and others (reviewed by D’Souza-Schorey and Chavrier 2006). Cx43 is known to be associated with different cancers (Mesnil 2002; Oliveira et al. 2005). Controversial data exist for various forms of the disease, e.g. cervical, prostate, SLC lung cancer, gliomas and melanomas, as to whether connexin expression (e.g. Cx43) is downregulated or whether e.g. turnover at the PM is increased (Herve et al. 2007). In summary, our results suggest that Arf6 in its active, GTP-bound form may carry important functions in directing quick disassembly of Cx43 plaques in developing and migrating cells. The fact that Cx43 in Arf6Q67L-transfected cells co-localized at the cell periphery with paxillin supports this possibility. It was shown that expression of Cx43 suppresses the growth of human glioblastoma cells (Huang et al. 2002). Why do cells reduce gap junction coupling during neoplastic transformation? Perhaps this is part of a protective mechanism against malignant transformation where cells may have tendency to limit the spread of apoptotic signaling, e.g. Ca2+-mediated waves or other signals that can easily pass through gap junctions. Thus, investigating the Arf6-dependence of gap junction communication is important in the context of understanding the loss of gap junction coupling between cancer cells.
Live cell data presented here provide a framework to explain how extracellular factors acting via small GTPases of the secretory pathway may regulate connexin transport to the cell surface. Interestingly, recent work shows the importance of intracellular trafficking and stabilization of Cx43 at the plasma membrane in astrocytes for the survival of dopaminergic (DA) neurons in a rotenone-induced model of Parkinson disease (Kawasaki et al. 2009). Gap junction levels are known to be increased during embryonic development. Similar regulation of cell–cell coupling by small GTPases is likely to be utilized during brain development when increased formation of specific neuronal circuits requires not a bulk flow of connexin molecules to the PM but specific regulation and fine tuning of their PM delivery. Gap junctions provide a platform for cell–cell communications in neuroepithelium elaborating formation of neuronal circuits in cerebral cortex to serve higher cognitive functions. Gap junction coupling was shown to regulate proliferation of neural progenitors during migration and the differentiation of young neurons in the embryonic cerebral cortex, acting both in the classical manner by coupling neural progenitors and through hemichannels that mediate the spread of calcium waves across cell populations in migrating neurons (for review see Elias and Kriegstein 2008). Our data thus address regulatory transport mechanisms underpinning cell–cell communication in tissues and organs. A more detailed understanding is important for the newly emerging field of regenerative medicine that aims to provide new tools in cell-based therapies.
Abbreviations
- ARF1:
-
ADP-ribosylation factor-1
- Cx43:
-
Connexin-43
- ER:
-
Endoplasmic reticulum
- ERES:
-
ER exit sites
- FLIM:
-
Fluorescence lifetime imaging microscopy
- FP:
-
Fluorescent protein
- FRET:
-
Fluorescence resonance energy transfer
- IP3:
-
Inositol 1,4,5-trisphosphate
- ns:
-
Nanoseconds
- PIP2:
-
Phosphatidylinositol (4,5)-bisphosphate
- PM:
-
Plasma membrane
References
Aridor M, Bannykh SI, Rowe T, Balch WE (1995) Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J Cell Biol 131:875–893
Baxter HC, Liu WL, Forster JL, Aitken A, Fraser JR (2002) Immunolocalisation of 14-3-3 isoforms in normal and scrapie-infected murine brain. Neuroscience 109:5–14
Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116:153–166
Bozhkova VP, Kvavilashvili IS, Rott NN, Chailakhyan LM (1974) Measurement of electrical coupling between cells of axolotl embryos during cleavage divisions. Sov J Dev Biol 4:480–482
Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB (1997) Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol 139:469–484
Butkevich E (2004) PhD thesis. http://webdoc.sub.gwdg.de/diss/2004/butkevich/butkevich.pdf
Butkevich E, Hulsmann S, Wenzel D, Shirao T, Duden R, Majoul I (2004) Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr Biol 14:650–658
Chailakhyan LM (1976) Contact membranes in cell cultures and embryonic tissues. Stud Biophys (Berlin) 56:23–24
Chailakhyan LM (1990) Ligand–receptor and junction-mediated cell–cell interactions: comparison of the two principles. Differentiation 45:1–6
Coblitz B, Shikano S, Wu M, Gabelli SB, Cockrell LM, Spieker M, Hanyu Y, Fu H, Amzel LM, Li M (2005) C-terminal recognition by 14-3-3 proteins for surface expression of membrane receptors. J Biol Chem 280:36263–36272
Couve A, Kittler JT, Uren JM, Calver AR, Pangalos MN, Walsh FS, Moss SJ (2001) Association of GABA(B) receptors and members of the 14-3-3 family of signaling proteins. Mol Cell Neurosci 17:317–328
D’Souza-Schorey C, Chavrier P (2006) ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7:347–358
Darrow BJ, Fast VG, Kléber AG, Beyer EC, Saffitz JE (1996) Functional and structural assessment of intercellular communication. Increased conduction velocity and enhanced connexin expression in dibutyryl cAMP-treated cultured cardiac myocytes. Circ Res 79:174–183
Dascher C, Balch WE (1994) Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem 269:1437–1448
Duden R (2003) ER-to-Golgi transport: COP I and COP II function (Review). Mol Membr Biol 20:197–207
Elias LA, Kriegstein AR (2008) Gap junctions: multifaceted regulators of embryonic cortical development. Trends Neurosci 31:243–250
Frigerio G, Grimsey N, Dale M, Majoul I, Duden R (2007) Two human ARFGAPs associated with COP-I-coated vesicles. Traffic 8:1644–1655
Fu H, Subramanian RR, Masters SC (2000) 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol 40:617–647
Giepmans BG, Verlaan I, Hengeveld T, Janssen H, Calafat J, Falk MM, Moolenaar WH (2001) Gap junction protein connexin-43 interacts directly with microtubules. Curr Biol 11:1364–1368
Goldberg GS, Lampe PD, Nicholson BJ (1999) Selective transfer of endogenous metabolites through gap junctions composed of different connexins. Nat Cell Biol 1:457–459
Herve JC, Bourmeyster N, Sarrouilhe D, Duffy HS (2007) Gap junctional complexes: from partners to functions. Prog Biophys Mol Biol 94:29–65
Huang R, Lin Y, Wang CC, Gano J, Lin B, Shi Q, Boynton A, Burke J, Huang RP (2002) Connexin 43 suppresses human glioblastoma cell growth by down-regulation of monocyte chemotactic protein 1, as discovered using protein array technology. Cancer Res 62:2806–2812
Jovanovic OF, Brown D, Donaldson JG (2006) An effector domain mutant of Arf6 implicates phospholipase D in endosomal membrane recycling. Mol Biol Cell 17:327–335
Kawasaki A, Hayashi T, Nakachi K, Nakachi K, Trosko JE, Sugihara K, Kotake Y, Ohta S (2009) Modulation of connexin 43 in rotenone-induced model of Parkinson’s disease. Neuroscience 160:61–68
Klopfenstein DR, Tomishige M, Stuurman N, Vale RD (2002) Role of phosphatidylinositol (4,5) bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell 109:347–358
Krutovskikh V (2002) Implication of direct host–tumor intercellular interactions in non-immune host resistance to neoplastic growth. Sem Cancer Biol 12:267–276
Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE (1994) Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol 125:51–65
Kumar NM, Gilula NB (1996) The gap junction communication channel. Cell 84:381–388
Li X, Su V, Kurata WE, Jin C, Lau AF (2008) A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J Biol Chem 283:5748–5759
Loewenstein WR (1981) Junctional intercellular communication: the cell-to-cell membrane channel. Physiol Rev 61:829–913
Lowe M, Kreis TE (1996) In vivo assembly of coatomer, the COP-I coat precursor. J Biol Chem 271:30725–30730
Majoul IV, Bastiaens PI, Soling HD (1996) Transport of an external Lys-Asp-Glu-Leu (KDEL) protein from the plasma membrane to the endoplasmic reticulum: studies with cholera toxin in Vero cells. J Cell Biol 133:777–789
Majoul I, Jia Y, Duden R (2006) In: Pawley JB (ed) Handbook of biological confocal microscopy, 3rd edn. Springer, Berlin, pp 788–808
Margeta-Mitrovic M (2002) Assembly-dependent assays in the detection of receptor–receptor interactions. Methods 4:311–317
Margeta-Mitrovic M, Jan YN, Jan LY (2000) A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27:97–106
Maza J, Das Sarma J, Koval M (2005) Defining a minimal motif required to prevent connexin oligomerization in the endoplasmic reticulum. J Biol Chem 280:21115–21121
Mesnil M (2002) Connexins and cancer. Biol Cell 94:493–500
Michelsen K, Schmid V, Metz J, Heusser K, Liebel U, Schwede T, Spang A, Schwappach B (2007) Novel cargo-binding site in the beta and delta subunits of coatomer. J Cell Biol 179:209–217
Miyawaki A, Tsien RY (2000) Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol 327:472–500
Musil LS, Goodenough DA (1990) Gap junctional intercellular communication and the regulation of connexin expression and function. Curr Opin Cell Biol 2:875–880
Naslavsky N, Weigert R, Donaldson JG (2004) Characterization of a nonclathrin endocytic pathway: membrane cargo and lipid requirements. Mol Biol Cell 15:3542–3552
O’Kelly I, Butler MH, Zilberberg N, Goldstein SA (2002) Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell 111:577–588
Oliveira R, Christov C, Guillamo JS, de Boüard S, Palfi S, Venance L, Tardy M, Peschanski M (2005) Contribution of gap junctional communication between tumor cells and astroglia to the invasion of the brain parenchyma by human glioblastomas. BMC Cell Biol 6:7. doi:10.1186/1471-2121-6-7
Paleotti O, Macia E, Luton F, Klein S, Partisani M, Chardin P, Kirchhausen T, Franco M (2005) The small G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J Biol Chem 280:21661–21666
Park DJ, Freitas TA, Wallick CJ, Guyette CV, Warn-Cramer BJ (2006) Molecular dynamics and in vitro analysis of Connexin43: a new 14-3-3 mode-1 interacting protein. Protein Sci 15:2344–2355
Rajan S, Preisig-Muller R, Wischmeyer E, Nehring R, Hanley PJ, Renigunta V, Musset B, Schlichthörl G, Derst C, Karschin A, Daut J (2002) Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3. J Physiol 545:13–26
Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci 21:3063–3072
Sohl G, Odermatt B, Maxeiner S, Degen J, Willecke K (2004) New insights into the expression and function of neural connexins with transgenic mouse mutants. Brain Res Rev 47:245–259
Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ (2000) PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28:887–898
Thomas T, Jordan K, Simek J, Shao Q, Jedeszko C, Walton P, Laird DW (2005) Mechanisms of Cx43 and Cx26 transport to the plasma membrane and gap junction regeneration. J Cell Sci 118:4451–4462
Tokuyasu KT (1978) A study of positive staining of ultrathin frozen sections. J Ultrastruct Res 63:287–307
Toyooka K, Muratake T, Watanabe H, Hayashi S, Ichikawa T, Usui H, Washiyama K, Kumanishi T, Takahashi Y (2002) Isolation and structure of the mouse 14-3-3 eta chain gene and the distribution of 14-3-3 eta mRNA in the mouse brain. Brain Res Mol Brain Res 100:13–20
Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67:509–544
Unger VM, Kumar NM, Gilula NB, Yeager M (1999) Three-dimensional structure of a recombinant gap junction membrane channel. Science 283:1176–1180
Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA (1999) Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell 10:4107–4120
VanSlyke JK, Naus CC, Musil LS (2009) Conformational maturation and post-ER multisubunit assembly of gap junction proteins. Mol Biol Cell 20:2451–2463
Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Güldenagel M, Deutsch U, Söhl G (2002) Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem 383:725–737
Xia H, Hornby ZD, Malenka RC (2001) An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology 41:714–723
Yang F, Moss LG, Phillips GN Jr (1996) The molecular structure of green fluorescent protein. Nat Biotech 14:1246–1251
Yuan H, Michelsen K, Schwappach B (2003) 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr Biol 13:638–646
Zerangue N, Schwappach B, Jan YN, Jan LY (1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22:537–548
Acknowledgments
We are grateful to Dr. J. Donaldson providing ARF6 constructs, Dr. B. Balch for Sar1 constructs, Dr. P. Luzio for the Lgp120 clone, to the Lambert instrument group and Dr. R. Hut for assistance with the FLIM measurements. We thank A. Helenius for helpful suggestions and comments on an early version of this manuscript. This work was supported by The Wellcome Trust (“Senior Research Fellowship”; grant 047578 to RD) and the Deutsche Forschungsgemeinschaft (Excellence Cluster “Inflammation at Interfaces”).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Majoul, I.V., Onichtchouk, D., Butkevich, E. et al. Limiting transport steps and novel interactions of Connexin-43 along the secretory pathway. Histochem Cell Biol 132, 263–280 (2009). https://doi.org/10.1007/s00418-009-0617-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-009-0617-x