Abstract
During endochondral ossification hypertrophic chondrocytes in the growth plate of fetal long bones, ribs and vertebrae play a key role in preparing growth plate cartilage for replacement by bone. In order to establish a reporter gene mouse to facilitate functional analysis of genes expressed in hypertrophic chondrocytes in this process, Col10a1- BAC reporter gene mouse lines were established expressing LacZ specifically in hypertrophic cartilage under the control of the complete Col10a1 gene. For this purpose, a bacterial artificial chromosome (BAC RP23-192A7) containing the entire murine Col10a1 gene together with 200 kb flanking sequences was modified by inserting a LacZ-Neo cassette into the second exon of Col10a1 by homologous recombination in E. coli. Transgenic mice containing between one and seven transgene copies were generated by injection of the purified BAC-Col10a1- lLacZ DNA. X-gal staining of newborns and embryos revealed strong and robust LacZ activity exclusively in hypertrophic cartilage of the fetal and neonatal skeleton of the transgenic offspring. This indicates that expression of the reporter gene in its proper genomic context in the BAC Col10a1 environment is independent of the integration site and reflects authentic Col10a1 expression in vivo. The Col10a1 specific BAC recombination vector described here will enable the specific analysis of effector gene functions in hypertrophic cartilage during skeletal development, endochondral ossification, and fracture callus healing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cartilage–bone transition by endochondral ossification is a highly complex process which determines not only longitudinal growth of long bones, ribs and vertebrae, but also plays a critical role in bone fracture callus healing, osteophyte formation and cartilage tissue engineering. Decisive steps in this process are proliferation, maturation and hypertrophy of chondrocytes in the growth plate which play a key role in mineralization, apoptosis and induction of resorption of hypertrophic cartilage by invading bone marrow. These events are regulated by synergistic action of several signaling pathways and transcriptions factors controlled by a multitude of growth factors and hormones.
A major signaling system regulating chondrocyte proliferation and maturation is controlled by PTHrP, which is induced by Indian hedgehog, stimulates chondrocyte proliferation and delays maturation through the PTHrP receptor PTHR1 [for reviews see (Kronenberg et al. 1998; Vortkamp 2000)]. Also FGF factors stimulate proliferation and block maturation through FGF receptor 3, but their action in the growth plate is more complex with respect to the differential distribution of different FGFs and FGF receptors in the various zones of the growth plate (Ornitz and Marie 2002).
The role of BMPs during chondrocyte maturation and hypertrophy is somewhat controversially discussed: several studies show that BMP factors such as BMP-6 and BMP-7 stimulate hypertrophic differentiation of chondrocytes and promote collagen X expression, thus preparing growth plate cartilage for replacement by endochondral bone (Enomoto-Iwamoto et al. 1998; Volk et al. 1998; Grimsrud et al. 2001). On the other hand, it was reported that BMP-2 and BMP-4 overexpression in developing chick limbs caused delayed hypertrophy of chondrocytes (Duprez et al. 1996).
The fourth family regulating chondrocyte maturation are Wnt factors and the Wnt/β-catenin signaling system (Dong et al. 2005), also acting in diverse directions. For example, Wnt 7a inhibits chondrogenesis and Wnt 5a impairs chondrocyte hypertrophy and type X collagen (Col10a1) expression in vitro (Daumer et al. 2004), but activation of the canonical β-catenin pathway by Wnt 8c and Wnt 9a induces chondrocytes maturation, hypertrophy and calcification, associated with upregulation of Col10a1 expression (Enomoto-Iwamoto et al. 2002; Dong et al. 2005, 2006; Hu et al. 2005).
Recently increasing evidence points to complex crosstalks between these signaling pathways. For example, there is experimental evidence for a coordination between Indian hedgehog and BMP signaling in chondrocyte proliferation (Minina et al. 2002). Targets of the signaling pathways in the growth plate are transcription factors such as Sox9 responsible for chondrogenic differentiation (Ng et al. 1997; Lefebvre and de Crombrugghe 1998) and cbfa1/runx2 inducing osteogenic differentiation (Karsenty et al. 1999; Ducy 2000) as well as chondrocyte maturation and Col10a1 expression (Kim et al. 1999; Enomoto et al. 2000; Zheng et al. 2003).
There is ample evidence in the literature that factors regulating chondrocyte maturation and hypertrophy may be also directly involved in the regulation of Col10a1 gene expression, e.g. PTHrP (Iwamoto et al. 1994; Ionescu et al. 2001; Riemer et al. 2002; Gebhard et al. 2004), Runx2 (Enomoto-Iwamoto et al. 2001; Leboy et al. 2001; Zheng et al. 2003), BMP2 (Volk et al. 1998; Grimsrud et al. 2001), or c-fos (Thomas et al. 2000; Riemer et al. 2002). Thus, in order to elucidate the exact role of the various signaling systems and involved factors regulating endochondral ossification, it would be useful to generate a reporter gene mouse in which the expression of LacZ is under the control of the complete Col10a1 gene regulatory sequences.
Previously we have reported on the generation of transgenic mouse lines in which LacZ expression is under the control of the 4.6 kb promoter of Col10a1 which includes a strong enhancer element (Gebhard et al. 2004) and runx2 binding sites (Zheng et al. 2003). We have shown that this promoter is sufficient to drive specific expression of LacZ reporter genes in hypertrophic chondrocytes of growth plate cartilage of long bones, ribs, vertebrae and sternal cartilage (Gebhard et al. 2004). Reporter gene expression was restricted to hypertrophic cartilage, but not all hypertrophic chondrocytes in the growth plate of 18.5-day embryos were found to be lacZ positive, and only weak lacZ expression was observed at early embryonic stages before day E17. Furthermore, not all transgenic founders containing the LacZ transgene showed lacZ activity in the growth plates, suggesting a high level of sensitivity to interfering activities of the genomic context of insertion or additional regulatory elements in the Col10a1 gene further upstream of the enhancer or downstream of the coding sequences.
Here we report on the construction of a BAC reporter mouse expressing lacZ under the control of the Col10a1 gene. A LacZ-Neo cassette was inserted into the second exon of Col10a1 within the context of a 215 kb BAC using a phage-based homologous recombination system in E. coli (Yu et al. 2000; Lee et al. 2001). Transgenic mouse lines established with this modified BAC show specific LacZlacZ expression at high levels in hypertrophic zones of long bones, ribs, vertebrae, mandibles and sterna of transgenic mouse lines. No significant unspecific expression was detected in other chondrogenic or non-chondrogenic tissues except some transient, probably unspecific X-gal reaction in the prenatal epidermis and hair papillae.
The robust and specific expression of Col10a1-based BAC recombineering vectors in transgenic mice opens new and unique possibilities to study the role of growth factors and transcription factors in chondrocyte hypertrophy and endochondral ossification, and to define further cis-acting regions in the Col10a1 gene.
Materials and methods
BAC clones
Murine Col10a1 containing BAC clones were selected from the UCSC Bioinformatic databases (http://www.genome.ucsc.edu/) (Kent et al. 2002); clone RP23-192A7 contained 211 kb of genomic DNA flanking the Col10a1 gene derived from the L129 mouse strain. RP23-192A7 (cloned in the pBACe3.6 vector) was obtained from the BACPAC Resources Center at Children’s Hospital Oakland Research Institute, (CHORI). The presence of the full-length Col10a1 gene was verified by PCR amplification using both primers for detecting the enhancer at −4.6 kb (Enmcol10a1-3′: ATT CTC GAG CTT TGG GAA GCA TGG TG ; Enmcol10a1-5′: CAA GAA ATC TTT GGG AAA TGA ATG AAT G ) (Gebhard et al. 2004) , and for a region 2036 bp downstream of the poly A site of the Col10a1 gene (mcol10a1-psg-5′: TCT ACC AGG AGG CCT CTC TTC AAT GTT AC; mcol10a1-psg-3′:
GGG AAT TCT TAC CTT AAA GTA GAT ACA TG (amplicon size 222 bp).
Construction of targeting vector placH+Col10a1-lacZ-frt-neo-frt
The 5′ recombination site was generated by PCR amplification of a 129 bp fragment mapping to the 113 bp 3′-end of intron 1 and 16 bp of the 5′-UTR encoded in exon 2 by using BAC RP23-192A7 as template (see Fig. 1). Terminal SalI and NcoI restriction sites were added by using primers Col10a1-5′SalI (5′-ACG CGT CGA CGA TAT CTC AGT CAT TTA AAA AAC CAT GA) and Col10a1-5′NcoI (5′-CAT GCC ATG GAT TTT CAG ATA GAT TCT GAA AAG CAG A. The product was digested with SalI and NcoI and cloned into the multiple cloning site 2 of plasmid placH (Zhou et al. 1995) kindly provided by Dr. V. Lefebvre, Cleveland) containing the lacZ gene and the 3′-end of the murine protamine 1 gene with an intron and poly A signal resulting in clone placH+5′COL10a1. Use of the NcoI site linked the lacZ reading frame to the start ATG of Col10a1.
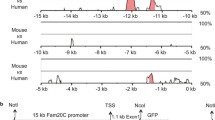
Generation of the BAC-Col10a1-lacZ-neo DNA for the transgenic expression of the lacZ reporter gene in hypertrophic cartilage. a Genomic structure of the murine Col10a1 gene, with exons (gray boxes) and introns. The sequences used for generation of the 5′ and 3′ homologous regions in exon 2 and intron 1 are shown in black. b The targeting vector placH- Col10a1-lacZ-frt-neo-frt contains Col10a1 derived 5′ and 3′ homology domains flanking a lacZ-frt-neo-frt cassette for homologous recombination in E. coli. The lacZ coding sequence was fused with the start ATG of the Col10a1 reading frame; shown is only the insert of the targeting vector, contained in the placH vector. c Partial map of the BAC-clone Col10a1-lacZ-frt-neo-frt clone after homologous recombination with the targeting vector placH-Col10a1-lacZ-frt-neo-frt
The 3′ recombination site was generated by overlapping primers corresponding to a 57 bp sequence of exon 2. Additional HindIII sites were introduced at both ends as well as a XhoI site at the 3′-end. The frt-neo-frt cassette of the pICGN21 vector (Yu et al. 2000; Lee et al. 2001); kindly provided by Dr. Copeland) harboring the neomycin phosphotransferase gene under the SV40 promoter, an HSV thymidine kinase and a poly A signal was amplified by PCR using primer FRTNeo5´HindIII (5′-AGT CAA GCT TTA ACT GAT CGC GGC CAG CTT GAA GTT) (pIGCN21 sequence pos. 4664–4689, marked in bold) and the primer Col10a1-3′HindIII (5′-TAA AGC TTC TCG AGT TGT GTC TTG GGG CTA GC AAG TGG GCC CTT TAT GCC TGT GGG CGT TTG GTA CCG TT C TAT TCC AGA AGT AGT GAG GAG GCT TT) and plasmid pICGN21 as template (pICGN2 sequences pos. 6103 to 6123 marked in italics) and Col10a1 from exon 2 (82 to 138, underlined). The product was digested with HindIII and inserted into the multiple cloning site behind the mP1 gene of placH+5′Col10a1. The resulting plasmid placH+Col10a1-lacZ-frt-neo-frt was verified to be correct by restriction digestion and sequencing for correct insertion and composition.
Homologous recombination and detection of BAC transgene
BAC RP23-192A7 was electroporated into the E. coli strain EL250 (DH10B[λcl857(cro-bioA)<>araC-PBAD flpe]) kindly provided by Dr. Neal G. Copeland, NCI, Frederick, MD, USA, containing a defective λ prophage which supplies essential functions (exo, bet, gam) to protect and enhance recombination of linear DNA (Lee et al. 2001).
The 5.1 kb recombination cassette was excised from plasmid placH+COL10a1-lacZ-frt-neo-frt by XhoI; complete linearization with XhoI was important to avoid amplification of the circular targeting vector in E. coli.
For homologous recombination 300 ng linear fragment was transformed by electroporation according to Dower (Dower et al. 1988) in 0.1 ml cuvettes containing 50 μl of ice-cold competent EL250 containing BAC RP23-192A7, using a Bio-Rad gene pulser set at 1,750 kV, 23 mF with a pulse controller set at 200 Ω. Electrocompetent bacteria were prepared as described in (Lee et al. 2001). A 15 min heat shock at 42°C was used for induction of the homologous recombination machinery according to (Yu et al. 2000; Lee et al. 2001). Transformed cells were incubated for 1.5 h at 32°C in 1 ml of LB and selected for chloramphenicol- and kanamycin-resistance. Resulting clones (RP23-192A7 + lacZ-frt-neo-frt) were screened for homologous recombination by PCR detecting the 5′ recombination event (amplicon 437 bp) using primers P1 (5′-TTT AGA GCA TTA TTT CAA GGC AGT TTC CA) and P2 (5′-CGG CAC CGC TTC TGG TGC CGG AAA CCA GGC), and the 3′ recombination event (product 378 bp) using primers P3 (5′-ACA GAA TAA AAC GCA CGG GTG TTG GGC GT) and P4 (5′-ATC ATT CCG CTG TAC TAG CTC AAG CCA ATC). Direct sequencing of BAC DNA was performed with primers P1 and P4.
BAC DNA purification and generation of transgenic mice
BAC-Col10a1 - lacZ-neo DNA from clone#11 was extracted from multiple E. coli EL250-BAC#11 minipreps by alkaline lysis (Sambrook et al. 2001), purified by potassium acetate precipitation, washed with ethanol and dissolved in Tris-EDTA (TE) buffer. For size and quality control, aliquots were subjected to pulsed field gel electrophoresis (PFGE) (Fig. 2). For purification, 50 μg BAC DNA of clone#11(BAC-Col10a1-LacZ-neo) were dissolved in TE buffer and concentrated to 500 μl by vacuum centrifugation. The DNA was linearized overnight with PISceI enzyme (NEBiolabs) cleaving the unique site in the BACe3.6 vector part. For chromatographic purification a 5 ml plastic pipette was packed with Sepharose CL-4B (Pharmacia) equilibrated in microinjection buffer (5 mM Tris/HCl, pH 7.4, 0.1 mM EDTA, 5 mM NaCl) (Zeilhofer et al. 2005), the linearized BAC#11-DNA was mixed with bromophenol blue and chromatographed. Half milliliter fractions were collected until the dye reached the column bottom, and BAC DNA concentration in each fraction was analyzed by PFGE using serial dilutions of BAC RP23-192A7 DNA as internal standard. The BAC DNA concentration of the main fraction was adjusted to 1.5 ng/μl, and NaCl concentration was increased to 100 mM to stabilize BAC DNA. Fractions were stored at 4°C until microinjection. Injection into fertilized oocytes and generation of transgenic mice was performed by standard techniques using oocytes from FVB mice and FVB/ FVBxC57BL6 F1 hybrids as described (Zeilhofer et al. 2005).
Characterization of one of the BAC-Col10a1-LacZ-frt-neo-frt clones (clone#11). Digestion with PISceI (lane 3), NotI (lane 4), PmeI (lane 5) and NruI (lane 6) resulted in fragments of the expected sizes. Sizes are compared with 1 kb marker (lane 1), 1 kb extension (lane 2) and PFG marker (lane 7), and the lengths are indicated. For microinjection, BAC #11 DNA was linearized with PISceI, purified by gel chromatography on Sepharose CL4B equilibrated in microinjection buffer and analyzed and quantitated by pulsed field gel electrophoresis
Genotyping of transgenic mouse lines
Transgenic founder animals were selected by PCR of tail biopsies using primers pairs P1/P2 and P3/P4 (see above) detecting recombination events. Each sample was standardized by PCR using P1/P4 primer pair which generates a 538 bp fragment from the wt Col10a1 allele. Founders derived from injections of F1 hybrids were crossed with C57BL6, whereas founders derived from FVB oocytes were crossed with FVB mice. For genotyping of newborn mice and embryos DNA was extracted from skin or placenta, and transgene DNA was identified by PCR as above.
Histological analysis and in situ hybridization
Newborns mice were anesthetized under CO2, dissected and subjected to X-gal staining, followed by either clearing with KOH fixation, embedding into paraffin or by cryotome sectioning as described (Zhou et al. 1995). Embryos at 12.5 to 16.5 day were left undissected and stained with X-gal. Paraffin and frozen sections were counterstained with eosin.
In situ hybridization for collagen X was performed as described previously (Aigner et al. 1992), but using a digoxigenin-labeled mouse Col10a1-specific anti-sense probe derived from the 3′-end of the Col10a1 gene instead of 32P-labeled dCTP. Non-radioactive in situ hybridization was done as described elsewhere (Schmidl et al. 2006), using alkaline phosphatase labeled anti-digoxigenin antibodies and BM purple (Roche) as color substrate according to the manufacturers protocol.
Determination of transgene copy number by Southern hybridization
For quantitative analysis of transgene copy number, genomic DNA was isolated from skin of newborns or from placenta of embryos and cleaved with BamHI. For standardization 5 μg wild-type DNA were mixed with 0.37 ng up to 18.38 ng RP23-192A7 + lacZ-frt-neo-frt DNA corresponding to 1 up to 50 copies of recombined BAC per lane. Samples for standardization and 5 μg of DNA from transgenic founders were cleaved with BamHI, separated on 1% agarose gels and blotted onto nylon membranes. Membranes were hybridized to with a 32P-labeled probe derived for from the pBACe3.6 vector, prepared by PCR using primers BAC11-5 (5′-TTTAAACGTGGCCAATATGGA) and BAC11-6 (5′-CGCGGATCCTCTCCCTAT). An 8 kb band was detected in transgenic samples and the intensity was measured in a FLA-3000 phosphoimager (Fujifilm) using the AIDA program (Advanced Image Data Analyzer, Raytest ).
Results
Construction of a placH-Col10a1-lacZ-frt-neo-frt targeting vector and homologous recombination
The BAC clone RP23-192A7 selected from the CHORI BAC library RPCI23 contains the mouse Col10a1 gene (7.2 kb) with 171 kb of upstream and 33 kb of downstream genomic regions. For insertion of the lacZ reporter gene into the second exon of the Col10a1 gene by homologous recombination, a targeting vector was constructed containing the lacZ gene linked to a neomycin resistance cassette flanked by two frt sites. The targeting vector was flanked by a 129 bp 5′-terminal homology arm overlapping parts of the 3′-end of intron 1 and 14 bp of exon 2 including the start ATG for fusion with NcoI. The 3′ recombination site mapped to a 57 bp sequence in the 3′ part of exon 2 (Fig. 1b). The vector construct was verified by restriction mapping, Southern blotting and sequencing. Initial attempts using shorter homology regions resulted in mostly incorrect recombination events.
For homologous recombination, the BAC clone RP23-192A7 was electroporated into the E. coli strain EL250, resulting in EL250/RP23-192A7. The Col10a1-lacZ-frt-neo-frt insert was excised from the targeting vector with XhoI (Fig. 1) and electroporated into electrocompetent EL250/RP23-192A7. Chloramphenicol- and kanamycin-resistant BAC-positive clones were selected and tested by PCR for homologous recombination by using two primer pairs P1/P2 and P3/P4 (see Fig. 1 and Materials and Methods) specific for the 5′- and 3′- recombination sites, respectively. To verify the integrity of the Col10a1 gene in its genomic context after homologous recombination, the presence of the enhancer at 4.6 kb upstream the transcription site and the 5′-end of a pseudogene downstream of the 3′ UTR sequences was verified by PCR with specific primers (see Fig. 1 and Materials and Methods) and sequencing of the BAC DNA. Furthermore, intact BAC ends were confirmed by PCR, using primers derived from the pBACe3.6 vector and the genomic insert.
To minimize BAC degradation, DNA was prepared from BAC-Col10a1-LacZ-Neo clone #11 by alkaline lysis and ethanol precipitation, avoiding absorption to minicolumns and shear stress. Control of the BAC DNA by PFGE confirmed the expected size of 220 kb (Fig. 2).
Generation of transgenic mice
DNA from BAC-Col10a1-lacZ-neo (clone #11) was linearized with PISceI at the unique restriction site in the pBACe3.6 vector sequence, purified by molecular sieve chromatography, and injected into the pronuclei of fertilized oocytes of FVB mice and FVB/C59Bl F1 hybrids. Both strains were by far superior in litter size and successful raising offspring as compared to C57/Bl6 mice. Out of 70 newborn pups, 13 were found harboring the lacZ gene after PCR analysis of genomic DNA, using primer pairs P1/P2 and P3/P4 (see Fig. 1). Southern blot analysis of genomic DNA (Fig. 3a) as well as Real- time PCR analysis of genomic DNA using Col10a1 intron specific primers showed that the founders contained between one and seven transgene copies (Table 1)
a Analysis of BAC transgene copy numbers in 13 transgenic founders by Southern hybridization. Genomic DNA was isolated from skin of newborns, cleaved with BamHI, separated on 1% agarose gels and blotted onto nylon membranes. Membranes were hybridized with a 586 bp 32P-labeled probe derived from the pBACe3.6 vector which recognizes an 8 kb band in transgenic samples. Band intensities were quantitated in a phosphoimager. Copy numbers of transgene BAC DNA (Table 1) were calculated on the basis of a standard curve established with serial dilutions of BAC DNA starting from 50 copies down to 1 copy of BAC RP23-.192A7, diluted in 5 µg of wt DNA. b Comparison of LacZ activity in the scapulae of three different BAC transgenic lines harboring different BAC copy numbers shows some correlation between staining intensity and BAC copy number. All specimen were from new born animals and stained under identical conditions with X-gal for 18 h
Seven of the 13 independent founder lines were analyzed so far for LacZ activity by X-gal staining of newborns or embryos (all F1 generation). All offspring analyzed revealed strong LacZ activity after X-gal staining which was restricted to the hypertrophic zones in all growth plates of long bones, ribs, vertebrae, sternebrae and to hypertrophic cartilage in the skull in E14.5- and E16.5-day embryos and newborns (see Figs. 4, 5, 6). The X-gal staining pattern was identical in all seven lines investigated, while staining intensity varied in relation to the BAC transgene copy number (Fig. 3b). Under identical staining conditions, X-gal staining was strongest in founder #1520 (seven transgene copies), followed by #800 (five copies) and #986 (three copies) (Fig. 3b).
Detection of lacZ expression in BAC-Col10a1-lacZ-Neo transgenic embryos by X-gal staining at E14.5 (a), E16.5 (b) and P1 (e) of the F1 generation of founder lines #1534 (a), #1504 (b), and #1520 (c). X-gal staining reveals strongest expression in chondrocytes in the hypertrophic zones of the scapula (s), humerus (h), femur (f), clavicula (cl) and all other long bones, furthermore in the hypertrophic zone of the ribs and in cervical vertebrae (c1, c2), (p pelvic bone). c, d In E16.5 embryos some X-gal staining was also seen in the epidermis of the tail (b arrow) and in hair papillae of the forehead and snout, and at nostrils (n). This expression is restricted to some basal keratinocytes in the epidermis, here shown in a section of the snout (d). It disappears after birth and may be a result of unspecific β-gal activity. No LacZ activity was detected in non-hypertrophic chondrocytes or in non-chondrogenic tissues.
X-gal staining of transgenic newborn mice of BAC-Col10a1-lacZ-neo (a, b founder line #800, F1; c–i founder line #1520, F1). All hypertrophic zones of the growth plates of long bones, ribs, and vertebrae exhibit strong LacZ activity. a Humerus, distal; b ulna, proximal; c forearm, d hand; e skull: C1 cervical vertebrae 1 (atlas); eb ecto-occipital bone; h hyoid bone; m mandibular condyle; MC Meckels cartilage; mh head of malleus; r head of ramus. f Thoracal vertebrae (left four) and cervical vertebrae (right three), view from ventral; g ribs; h spine, view form dorsal, i sternum
Paraffin sections of transgenic newborns (a–e) (founder line #1520, F1) and E16.5 day (g) (founder line #1534) embryos showing strong and specific LacZ activity in all hypertrophic chondrocytes, e.g. in the growth plate of the humerus (a) (distal), elbow joint of the radius (b), sternebrae (c), atlas (d), ribs (e) and spine (f). Comparison of the lacZ staining pattern in the elbow joint (h radius) with that observed after in situ hybridization with a Col10a1 probe (g) in a parallel section illustrates that LacZ is expressed in the BAC transgenic mice only in collagen X expressing hypertrophic chondrocytes
Specific expression of the BAC-Col10a1-LacZ-neo transgene in hypertrophic cartilage
Various stages of embryonic development were analyzed for expression of the BAC-Col10a1-lacZ-neo transgene by X-gal staining. In E14.5-day embryos, strong LacZ activity was seen in all ribs, the hypertrophic zones of scapula, humerus, radius and ulna in the forearm and femur, tibia and fibula of the hindleg, as well as in mandibles and cervical vertebrae (Fig. 4a). At day 16.5, the extent of X-gal staining expanded in the diaphyseal region of scapula, humerus and in the ribs, corresponding to the expansion of the hypertrophic zones at this embryonic stage. (Fig. 4b, e) Sectioning of X-gal-stained embryos revealed that the lacZ transgene expression in the skeleton occurred exclusively in the hypertrophic zone of growth plates and ribs (see Figs. 5, 6), correlating with collagen X expression as seen by in situ hybridization (Fig. 6g, h).
At embryonic stages E12.5 to 16.5 X-gal staining also occurred in some regions of the epidermis (Fig. 4b, arrow, Fig. 4c, d), mainly in the papillae of the forehead in the snout (Fig. 4c), and in the skin of the tail, hindlegs, forelegs, and shoulder It was restricted to the basal layer of keratinocytes (Fig. 4d) and seen at E12.5 day before the first appearance of hypertrophic chondrocytes in cartilage, but was not detected after birth. It seemed unspecific as no α1(X) mRNA signals were seen in the epidermis or hair papillae at any stage of development by in situ hybridization (N. Adam and K. von der Mark, unpublished data).
Transgenic newborn F1 offspring of all seven founders analyzed so far revealed an even and strong LacZ activity in all growth plates of long bones (Figs. 4e, 5a, b), ribs (Figs. 4e, 5g), vertebrae (Figs. 4e, 5f, h), and sternebrae (Figs. 4e, 5i). In the skull, LacZ activity was seen in mandibular condyles, in hyoid cartilage, heads of malleus and ramus, ectooccipital and basisphenoid bone anlagen, cervical vertebrae and other ossifying cartilage anlagen (Fig. 5e). The high intensity of X-gal staining allowed the visualization of discrete zones of Col10a1 expression in hypertrophic cartilage, for example the inner and outer aspects of the thoracal and cervical vertebrae (Fig. 5f, h).
Analysis of the X-gal staining pattern in paraffin and frozen sections of transgenic embryos and newborn mice confirmed that the LacZ activity was restricted to hypertrophic chondrocytes (Fig. 6), shown here for the humerus (a), radius (b), sternum (c), atlas (d), and ribs (e, f). The X-gal staining pattern correlated well with the expression pattern of α1(X) collagen mRNA seen by in situ hybridization analysis of a similar section (Fig. 6g, h). Neither non-hypertrophic chondrocytes nor any non-chondrocytic cell types revealed detectable levels of LacZ-activity in newborn transgenic animals. After prolonged staining, a non-specific reaction appeared in osteoclasts of long bones in transgenic and wildtype animals owing to endogenous galactosidase activity. Regardless of the transgene copy number, the LacZ activity in the offspring of all founders was seen persistently in all hypertrophic chondrocytes of the developing skeleton.
Discussion
In a previous report on the generation of Col10a1-specific reporter gene mouse lines the expression of lacZ under the control of a 4.6 kb mouse Col10a1 promoter region was restricted to hypertrophic cartilage, indicating that the -4.6 kb mouse promoter including a 500 bp enhancer element located at the 5′-end of this region is sufficient for tissue specific expression (Gebhard et al. 2004). Also in another transgenic mouse expressing LacZ under a −4.0 kb proximal Col10a1 promoter lacking the enhancer, lacZ expression was restricted to hypertrophic cartilage, although mostly to the lower hypertrophic zone (Zheng et al. 2003). The Col10a1 enhancer has been originally described in the human COL10A1 gene; it was located between −1.9 and −2.4 kb by in vitro transfection studies in primary hypertrophic chondrocytes with various reporter gene constructs (Thomas et al. 1995; Beier et al. 1997; Chambers et al. 2002; Riemer et al. 2002). This enhancer is active in hypertrophic, but not in resting zone chondrocytes. Further upstream, a silencer element between −2.4 and −2.8 kb suppressed transcription in non-chondrogenic cells (Beier et al. 1997). The enhancer element is highly conserved in mammals with 78–85% sequence identity between human, bovine and murine Col10a1 genes (Gebhard et al. 2004) and contains a conserved AP-1 site which is essential for high transcription of enhancer- reporter genes in hypertrophic chondrocytes (Gebhard et al. 2004). In the murine gene, the enhancer is located between −4.1 and −4.6 kb due to insertion of two virus-related sequences in the promoter (Gebhard et al. 2004). Further cis-regulatory elements in the 4.6 kb promoter include an AP-1 site in the proximal promoter (Harada et al. 1996) and several runx2 bindings sites (OSE elements), (Zheng et al. 2003).
Yet, although lacZ transgene expression in the 4.6 kb Col10a1–LacZ reporter mice was specific and restricted to hypertrophic chondrocytes, not all hypertrophic chondrocytes in the growth plates of 18.5-day embryos were found to be LacZ positive, and only weak LacZ expression was observed at early embryonic stages before day E17 (Gebhard et al. 2004). Furthermore, not all transgenic founders containing the lacZ transgene were stained with X-gal in the growth plates, suggesting positional effects of additional regulatory elements in the Col10a1 gene further upstream of the enhancer or downstream of the coding sequences. It may also indicate a high level of sensitivity to interfering activities of the genomic context of insertion. It is possible that the −4.6 kb promoter including the first intron contains essential silencing elements required for suppressing Col10a1 expression in non-hypertrophic cartilage and in non-chondrogenic tissues, but possibly lacks additional regulatory elements beyond the enhancer region.
In contrast, in the Col10a1-BAC transgenic lines presented here, all hypertrophic chondrocytes showed strong and even LacZ expression in all zones of endochondral ossification in embryonic and postnatal transgenic animals. The LacZ reaction of hypertrophic cartilage became visible already after 3–4 h of X-gal staining, while under the control of the 4.6 kb Col10a1 promoter significant LacZ activity in transgenic mice did not become visible before 18 h staining time (Gebhard et al. 2004).
Recently, evidence is accumulating that in many genes further tissue-specific enhancers are located in large distances from the start site of transcription. For example, regulatory sequences were identified 20–30 kb upstream of the transcription start site of the Myo D gene (Goldhamer et al. 1992) or the Ubx gene in Drosophila (Irvine et al. 1991), up to 270 kb downstream in the mouse BMP5 gene (DiLeone et al. 2000) or between 290K kb upstream and 450 kb downstream of the Sox9 gene (Bagheri-Fam et al. 2006). Thus we cannot exclude that additional tissue-specific regulatory elements may be present in the Col10a1 gene, either upstream in the region between −170 kb and the enhancer at −4.6 kb, or downstream of the 3′-end of the Col10a1 gene. The BAC transgenic reporter lines established here would facilitate the identification of such elements by generating further mutants using the homologous recombination technique (DiLeone et al. 2000).
Reasons for the high reporter gene expression levels in the BAC-Col10a1-lacZ-neo reporter lines in comparison to the 4.6 kb Col10a1-lacZ mice are for one part the high copy number of BAC transgenes, and for the other part the favorable und undisturbed conditions for transgene expression in the BAC environment. The X-gal staining intensity in founder line #1520 harboring seven BAC transgene copies was clearly higher than that of founder line #986 harboring only three BAC copies. But in addition, the use of targeted BAC recombination permits the expression of genes in their genomic context, containing most relevant regulatory regions in the correct orientation and distance, in contrast to conventional transgenic mice generated by injection of short reporter genes. This strategy is less sensitive to unpredictable influences by regions surrounding the random integration site (Mortlock et al. 2003). The expression pattern of LacZ, however, was very similar in all seven BAC lines analyzed so far, apparently independent of the integration sites and the copy number of the BAC transgene.
Most E14.5- to E16.5-day BAC-Col10a1-lacZ-neo transgenic embryos showed some X-gal staining in hair follicles of the forehead and snout, and of the epidermis of the tail, legs and shoulder, which disappears after birth. In the X-gal positive spots, staining was restricted to the basal layer of keratinocytes. Since these cells do not express type X collagen, the effect may be due to endogenous galactosidase activity, or a result of LacZ activation by other, Col10a1-unrelated regulatory elements located in the BAC clone.
In addition to Col10a1, the BAC clone RP23-192A7 contains coding regions of three additional genes Tspyl-1, Tspyl1-4 and Nt5dc1. Whether these genes are expressed under their endogenous promoters in the BAC transgenic mice is currently investigated. If they are expressed, they could affect the phenotype of the established transgenic mouse lines, but no phenotypic alteration in skeletal elements nor in other tissues have been observed in transgenic embryos, newborn or adults as compared to wildtype littermates.
The high intensity of X-gal staining under the control of the BAC-Col10a1 promoter allowed the visualization of discrete zones of Col10a1 expression in hypertrophic cartilage of anatomical structures not investigated so far in such detail and completeness, for example the skull or the inner and outer aspects of the thoracal and cervical vertebrae. The data presented in this study illustrate the superior potential of reporter gene expression analysis in combination with BAC recombineering technique as compared to conventional in situ hybridization analysis.
The robust and specific expression of the lacZ reporter gene under the control of BAC-Col10a1 environment furthermore shows that the placH+Col10a1-lacZ-frt-neo-frt targeting vector may be an extremely versatile vector for the targeted overexpression of other genes in hypertrophic chondrocytes of growth plates with high efficiency and specificity. Thus, the Col10a1-based BAC vector may be a useful tool to analyze the role of effector genes in skeletal development, endochondral ossification, and fracture callus formation.
References
Aigner T, Stoess H, Weseloh G, Zeiler G, von der Mark K (1992) Activation of collagen type II expression in osteoarthritic and rheumatoid cartilage. Virchows Archiv B Cell Pathol 62:337–345
Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G (2006) Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol 291:382–397
Beier F, Vornehm S, Pöschl E, von der Mark K, Lammi MJ (1997) Localization of silencer and enhancer elements in the human type X collagen gene. J Cell Biochem 66:210–218
Chambers D, Young DA, Howard C, Thomas JT, Boam DS, Grant ME, Wallis GA, Boot-Handford RP (2002) An enhancer complex confers both high-level and cell-specific expression of the human type X collagen gene. FEBS Lett 531:505–508
Daumer KM, Tufan AC, Tuan RS (2004) Long-term in vitro analysis of limb cartilage development: involvement of Wnt signaling. J Cell Biochem 93:526–541
DiLeone RJ, Marcus GA, Johnson MD, Kingsley DM (2000) Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc Natl Acad Sci USA 97:1612–1617
Dong Y, Drissi H, Chen M, Chen D, Zuscik MJ, Schwarz EM, O’Keefe RJ (2005) Wnt-mediated regulation of chondrocyte maturation: modulation by TGF-beta. J Cell Biochem 95:1057–1068
Dong YF, Soung do Y, Schwarz EM, O’Keefe RJ, Drissi H (2006) Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J Cell Physiol 208:77–86
Dower WJ, Miller JF, Ragsdale CW (1988) High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16:6127–6145
Ducy P (2000) Cbfa1: a molecular switch in osteoblast biology. Dev Dyn 219:461–471
Duprez D, Bell EJ, Richardson MK, Archer CW, Wolpert L, Brickell PM, Francis-West PH (1996) Overexpression of BMP-2 and BMP-4 alters the size and shape of developing skeletal elements in the chick limb. Mech Dev 57:145–157
Enomoto-Iwamoto M, Iwamoto M, Mukudai Y, Kawakami Y, Nohno T, Higuchi Y, Takemoto S, Ohuchi H, Noji S, Kurisu K (1998) Bone morphogenetic protein signaling is required for maintenance of differentiated phenotype, control of proliferation, and hypertrophy in chondrocytes. J Cell Biol 140:409–418
Enomoto-Iwamoto M, Enomoto H, Komori T, Iwamoto M (2001) Participation of Cbfa1 in regulation of chondrocyte maturation. Osteoarthr Cartil 9(Suppl A):S76–S84
Enomoto-Iwamoto M, Kitagaki J, Koyama E, Tamamura Y, Wu C, Kanatani N, Koike T, Okada H, Komori T, Yoneda T, Church V, Francis-West PH, Kurisu K, Nohno T, Pacifici M, Iwamoto M (2002) The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol 251:142–156
Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, Kishimoto T, Komori T (2000) Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem 275:8695–8702
Gebhard S, Poschl E, Riemer S, Bauer E, Hattori T, Eberspaecher H, Zhang Z, Lefebvre V, de Crombrugghe B, von der Mark K (2004) A highly conserved enhancer in mammalian type X collagen genes drives high levels of tissue-specific expression in hypertrophic cartilage in vitro and in vivo. Matrix Biol 23:309–322
Goldhamer DJ, Faerman A, Shani M, Emerson CP Jr (1992) Regulatory elements that control the lineage-specific expression of myoD. Science 256:538–542
Grimsrud CD, Romano PR, D’Souza M, Puzas JE, Schwarz EM, Reynolds PR, Roiser RN, O’Keefe RJ (2001) BMP signaling stimulates chondrocyte maturation and the expression of Indian hedgehog. J Orthop Res 19:18–25
Harada S, Sampath TK, Rodan GA (1996) Identification of an AP1-like response region for osteogenic protein-1 in type X collagen promoter. Annu Acad Sci NY 785:259–60:259–260
Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F (2005) Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 132:49–60
Ionescu AM, Schwarz EM, Vinson C, Puzas JE, Rosier R, Reynolds PR, O’Keefe RJ (2001) PTHrP modulates chondrocyte differentiation through AP-1 and CREB signaling. J Biol Chem 276:11639–11647
Irvine KD, Helfand SL, Hogness DS (1991) The large upstream control region of the Drosophila homeotic gene Ultrabithorax. Development 111:407–424
Iwamoto M, Jikko A, Murakami H, Shimazu A, Nakashima K, Takigawa M, Baba H, Suzuki I, Kato Y (1994) Changes in parathyroid hormone receptors during chondrocyte differentiation. J Biol Chem 269:17245–17251
Karsenty G, Ducy P, Starbuck M, Priemel M, Shen J, Geoffroy V, Amling M (1999) Cbfa1 as a regulator of osteoblast differentiation and function. Bone 25:107–108
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Kim IS, Otto F, Zabel B, Mundlos S (1999) Regulation of chondrocyte differentiation by Cbfa1. Mech Dev 80:159–170
Kronenberg HM, Lanske B, Kovacs CS, Chung UI, Lee K, Segre GV, Schipani E, Juppner H (1998) Functional analysis of the PTH/PTHrP network of ligands and receptors. Recent Prog Horm Res 53:283–301; discussion 283–301; discussion 301–
Leboy P, Grasso-Knight G, D’Angelo M, Volk SW, Lian JV, Drissi H, Stein GS, Adams SL (2001) Smad–Runx interactions during chondrocyte maturation. J Bone Joint Surg Am 83-A(Suppl 1):S15–S22
Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65
Lefebvre V, de Crombrugghe B (1998) Toward understanding SOX9 function in chondrocyte differentiation. Matrix Biol 16:529–540
Minina E, Kreschel C, Naski MC, Ornitz DM, Vortkamp A (2002) Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev Cell 3:439–449
Mortlock DP, Guenther C, Kingsley DM (2003) A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res 13:2069–2081
Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P (1997) SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol 183:108–121
Ornitz DM, Marie PJ (2002) FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev 16:1446–1465
Riemer S, Gebhard S, Beier F, Poschl E, von der Mark K (2002) Role of c-fos in the regulation of type X collagen gene expression by PTH and PTHrP: localization of a PTH/PTHrP-responsive region in the human COL10A1 enhancer. J Cell Biochem 86:688–699
Sambrook J, MacCallum P, Russell D (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schmidl M, Adam N, Surmann-Schmitt C, Hattori T, Stock M, Dietz U, Decrombrugghe B, Poschl E, von der Mark KC (2006) Twisted gastrulation modulates BMP- induced collagen II and X expression in chondrocytes in vitro and in vivo. J Biol Chem 281 (in press)
Thomas JT, Sweetman WA, Cresswell CJ, Wallis GA, Grant ME, Boot-Handford RP (1995) Sequence comparison of three mammalian type-X collagen promoters and preliminary functional analysis of the human promoter. Gene 160:291–296
Thomas DP, Sunters A, Gentry A, Grigoriadis AE (2000) Inhibition of chondrocyte differentiation in vitro by constitutive and inducible overexpression of the c-fos proto-oncogene. J Cell Sci 113:439–450
Volk SW, Luvalle P, Leask T, Leboy PS (1998) A BMP responsive transcriptional region in the chicken type X collagen gene. J Bone Miner Res 13:1521–1529
Vortkamp A (2000) The Indian hedgehog–PTHrP system in bone development. Ernst Schering Res Found Workshop:191–209
Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97:5978–5983
Zeilhofer HU, Studler B, Arabadzisz D, Schweizer C, Ahmadi S, Layh B, Bosl MR, Fritschy JM (2005) Glycinergic neurons expressing enhanced green fluorescent protein in bacterial artificial chromosome transgenic mice. J Comp Neurol 482:123–141
Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B (2003) Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol 162:833–842
Zhou G, Garofalo S, Mukhopadhyay K, Lefebvre V, Smith CN, Eberspaecher H, de Crombrugghe B (1995) A 182 bp fragment of the mouse pro alpha 1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J.Cell Sci 108:3677–3684
Acknowledgments
We wish to express our sincere thanks to Drs. N.A. Copeland and E. Chiang Lee, NCI, Fredericks, MD, for providing the E. coli strain EL250 and the piGCN21 vector, and to Drs. Hsin-Fang Yang-Yen, IMB, Academia Sinica, Taipei and R.Behringer, MD Anderson Cancer Center, Houston for their valuable advice in BAC recombination methods. We gratefully acknowledge the generous support of Dr. Reinhard Fässler, Max-Planck-Institute of Biochemistry, Martinsried, Germany, of the mouse work done at the Max-Planck Institute of Biochemistry and to valuable advice by T. Samson, Department of Experimental Medicine I, Erlangen. The work was financially supported by a grant from the Deutsche Forschungsgemeinschaft to K.v.d.M. (MA534-18) and by a NIH grant 1P01 AR42919 to B.deC..
Author information
Authors and Affiliations
Corresponding author
Additional information
Sonja Gebhard and Takako Hattori equally contributed to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Gebhard, S., Hattori, T., Bauer, E. et al. BAC constructs in transgenic reporter mouse lines control efficient and specific LacZ expression in hypertrophic chondrocytes under the complete Col10a1 promoter. Histochem Cell Biol 127, 183–194 (2007). https://doi.org/10.1007/s00418-006-0236-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-006-0236-8