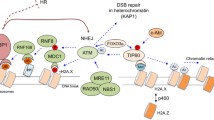
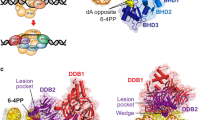
Abstract
The structure of chromatin is the major factor determining the rate and efficiency of DNA repair. Chromatin remodeling events such as rearrangement of nucleosomes and higher order chromatin structures are indispensable features of repair processes. During the last decade numerous chromatin proteins have been identified that preferentially bind to different types of DNA damage. The HMGB proteins, which preferentially interact with DNA intrastrand crosslinks induced by cisplatin, are the archetypal example of such proteins. Several hypothetical models have been proposed describing the role of such damage-binding chromatin proteins. The damage shielding model postulates that binding of chromatin proteins to damaged DNA might disturb damage recognition by repair factors and impair its removal. Alternatively, the damage-recognition/signaling model proposes that the binding of specific chromatin proteins to damaged DNA could serve as a hallmark to be recognized by repair proteins. Additionally, the binding of specific chromatin proteins to damaged DNA could induce chromatin remodeling at the damage site and indirectly affect its repair. This paper aims to critically review current experimental data in relation to such possible roles of chromatin proteins.


Similar content being viewed by others
References
Agresti A, Bianchi ME (2003) HMGB proteins and gene expression. Curr Opin Genet Dev 13:170–178
van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788
Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW (2002) Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A 99:8173–8178
Bellon SF, Coleman JH, Lippard SJ (1991) DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry 30:8026–8035
Birger Y, West KL, Postnikov YV, Lim Y-H, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M (2003) Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J 22:1665–1675
Brown SJ, Kellett PJ, Lippard SJ (1993) Ixr1, a yeast protein that binds to platinated DNA and confers sensitivity to cisplatin. Science 261:603–605
Bruhn SL, Pil PM, Essigmann JM, Housman DE, Lippard SJ (1992) Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc Natl Acad Sci USA 89:2307–2311
Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276:42462–42467
Bustin M (2001a) Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem Sci 26:152–153
Bustin M (2001b) Chromatin unfolding and activation by HMGN* chromosomal proteins. Trends Biochem Sci 26:431–437
Bustin M, Reeves R (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol 54:35–100
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A (2002) Genomic instability in mice lacking histone H2AX. Science 296:922–927
Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5:675–679
Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A (2000) Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science 290:1962–1965
Chu G (1994) Cellular responses to cisplatin. The roles of DNA-binding proteins and DNA repair. J Biol Chem 269:787–790
Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P (2001) The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat Res 486:89–97
Felsenfeld G, Groudine M (2003) Controlling the double helix. Nature 421:448–453
Fousteri M, van Hoffen A, Vargova H, Mullenders LHF (2005) Repair of DNA lesions in chromosomal DNA. Impact of chromatin structure and Cockayne syndrome proteins. DNA Repair 4:919–925
Fyodorov DV, Kadonaga JT (2001) The many faces of chromatin remodeling: switching beyond transcription. Cell 106:523–525
Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G (1996) Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86:887–896
Gong F, Kwon Y-H, Smerdon MJ (2005) Nucleotide excision repair in chromatin and the right of entry. DNA Repair 4:884–896
He Q, Liang CH, Lippard SJ (2000) Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc Natl Acad Sci USA 97:5768–5772
Hefferin ML, Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4:639–648
Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A (1994) HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci USA 91:10394–10398
Hughes EN, Engelsberg BN, Billings PC (1992) Purification of nuclear proteins that bind to cisplatin-damaged DNA; identity with high mobility group proteins 1 and 2. J Biol Chem 267:13520–13527
Iizuka M, Smith MM (2003) Functional consequences of histone modifications. Curr Opin Gen Dev 13:154–160
Imamura T, Izumi H, Nagadani G, Ise T, Nomoto M, Iwamoto Y, Kohno K (2001) Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein. J Biol Chem 276:7534–7540
Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamoto T, Ohga T, Uchiumi T, Kuwano M, Kohno K (1999) Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res 59:342–346
Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ (2003) Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol 160:1017–1027
Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K (2002) NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 12:1846–1851
Kunz C, Zurbriggen K, Fleck O (2003) Mutagenesis of the HMGB (high-mobility group B) protein Cmb1 (cytosine-mismatch binding 1) of Schizosaccharomyces pombe: effects on recognition of DNA mismatches and damage. Biochem J 372:651–660
Landsman D, Bustin M (1993) A signature for the HMG-1 box DNA-binding proteins. Bioessays 15:539–546
Lanuszewska J, Widlak P (2000) High mobility group 1 and 2 proteins bind preferentially to DNA that contains bulky adducts induced by benzo(a)pyrene diol epoxide and N-acetoxy-acetylaminofluorene. Cancer Lett 158:17–25
Lindsey GG, Orgeig S, Thompson P, Davies N, Maeder DL (1991) Extended C-terminal tail of wheat histone H2A interacts with DNA of the “linker” region. J Mol Biol 218:805–813
Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J (2004) Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 23:2674–2683
Malina J, Kasparkova J, Natile G, Brabec V (2002) Recognition of major DNA adducts of enantiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem Biol 9:629–638
Marchenko ND, Zaika A, Moll UM (2000) Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 275:16202–16212
McA’Nulty MM, Whitehead JP, Lippard SJ (1996) Binding of Ixr1, a yeast HMG-domain protein, to cisplatin-DNA adducts in vitro and in vivo. Biochemistry 35:6089–6099
Meijer M, Smerdon MJ (1999) Accessing DNA damage in chromatin: insights from transcription. BioEssays 21:596–603
Moggs JG, Almouzni G (1999) Chromatin rearrangements during nucleotide excision repair. Biochimie 81:45–52
Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X (2004) INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775
Nagaki S, Yamamoto M, Yumoto Y, Shirakawa H, Yoshida M, Teraoka H (1998) Non-histone chromosomal proteins HMG1 and 2 enhance ligation reaction of DNA double-strand breaks. Biochem Biophys Res Commun 246:137–141
Nagatani G, Nomoto M, Takano H, Ise T, Kato K, Imamura T, Izumi H, Makishima K, Kohno K (2001) Transcriptional activation of the human HMG1 gene in cisplatin-resistant human cancer cells. Cancer Res 61:1592–1597
Narlikar GJ, Fan H-Y, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487
Ohndorf UM, Whitehead JP, Raju NL, Lippard SJ (1997) Binding of tsHMG, a mouse testis-specific HMG-domain protein, to cisplatin-DNA adducts. Biochemistry 36:14807–14815
Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399:708–712
Pasheva EA, Pashev IG, Favre A (1998) Preferential binding of high mobility group 1 protein to UV-damaged DNA. Role of the COOH-terminal domain. J Biol Chem 273:24730–24736
Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895
Peterson CL, Cote J (2004) Cellular machineries for chromosomal DNA repair. Genes Dev 18:602–616
Peterson CL, Laniel M-A (2004) Histones and histone modifications. Curr Biol 14:R546–R551
Pil PM, Lippard SJ (1992) Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science 256:234–237
Reed SH (2005) Nucleotide excision repair in chromatin: the shape of things to come. DNA Repair 4:909–918
Reeves R, Adair JE (2005) Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair 4:926–938
Rich T, Allen RL, Wyllie AH (2000) Defying death after DNA damage. Nature 407:777–783
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868
Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146:905–916
Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM (2000) Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem 275:9390–9395
Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85
Siddik ZH (2003) Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22:7265–7279
Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA (2004) ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 64:2390–2396
Taneja N, Davis M, Choy JS, Beckett MA, Singh R, Kron SJ, Weichselbaum RR (2004) Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J Biol Chem 279:2273–2280
Thoma F (1999) Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J 18:6585–6598
Thoma F (2005) Repair of UV lesions in nucleosomes—intrinsic properties and remodeling. DNA Repair 4:855–869
Thomas JO, Travers AA (2001) HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26:167–174
Treiber DK, Zhai X, Jantzen HM, Essigmann JM (1994) Cisplatin-DNA adducts are molecular decoys for the ribosomal RNA transcription factor hUBF (human upstream binding factor). Proc Natl Acad Sci USA 91:5672–5676
Trimmer EE, Zamble DB, Lippard SJ, Essigmann JM (1998) Human testis-determining factor SRY binds to the major DNA adduct of cisplatin and a putative target sequence with comparable affinities. Biochemistry 37:352–362
Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555–560
Vichi P, Coin F, Renaud JP, Vermeulen W, Hoeijmakers JH, Moras D, Egly JM (1997) Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J 16:7444–7456
Wang JF, Bashir M, Engelsberg BN, Witmer C, Rozmiarek H, Billings PC (1997) High mobility group proteins 1 and 2 recognize chromium-damaged DNA. Carcinogenesis 18:371–375
Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14:927–939
Ward IM, Minn K, Jorda KG, Chen J (2003) Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem 278:19579–19582
Wei M, Burenkova O, Lippard SJ (2003) Cisplatin sensitivity in Hmgb1−/− and Hmgb1+/+ mouse cells. J Biol Chem 278:1769–1773
Wong B, Masse JE, Yen YM, Giannikopoulos P, Feigon J, Johnson RC (2002) Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein Nhp6A. Biochemistry 41:5404–5414
Yaneva J, Leuba SH, van Holde K, Zlatanova J (1997) The major chromatin protein histone H1 binds preferentially to cis-platinum-damaged DNA. Proc Natl Acad Sci USA 94:13448–13451
Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, Kasai H, Kohno K (2002) Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem Biophys Res Commun 295:945–951
Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K (2003) p53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res 63:3729–3734
Yuan F, Gu L, Guo S, Wang C, Li G-M (2004) Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J Biol Chem 279:20935–20940
Zamble DB, Jacks T, Lippard SJ (1998) p53-dependent and -independent responses to cisplatin in mouse testicular teratocarcinoma cells. Proc Natl Acad Sci USA 95:6163–6168
Zamble DB, Mikata Y, Eng CH, Sandman KE, Lippard SJ (2002) Testis-specific HMG-domain protein alters the responses of cells to cisplatin. J Inorg Biochem 91:451–462
Zhai X, Beckmann H, Jantzen HM, Essigmann JM (1998) Cisplatin-DNA adducts inhibit ribosomal RNA synthesis by hijacking the transcription factor human upstream binding factor. Biochemistry 37:16307–16315
Acknowledgements
The authors wish to thank Michel Bustin for a human recombinant HMGB1, Kimitoshi Kohno for a human recombinant mtTFA and William T. Garrard for help in preparation of the manuscript. This work was supported by the Polish Ministry of Science and Informatics, Grant KBN 4T11F01824.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Widlak, P., Pietrowska, M. & Lanuszewska, J. The role of chromatin proteins in DNA damage recognition and repair Mini-review . Histochem Cell Biol 125, 119–126 (2006). https://doi.org/10.1007/s00418-005-0053-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-005-0053-5