Abstract
Purpose
Primary cancers of the eye are common in ocular diseases. The objective of this study was to explore the underlying mechanisms and the potential target genes in multiple ocular cancers by bioinformatics approach.
Method
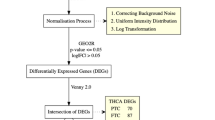
These gene expression profiles of GSE24673 (Retinoblastoma), GSE44295 (Uveal melanoma), and GSE103439 (Basal cell carcinoma of the eyelid) were downloaded from Gene Expression Omniniub (GEO) database. The differentially expressed genes (DEGs) in the three gene chips were identified by limma package in R software and gene integration was performed by using “RobustRankAggreg” package. Gene set enrichment analysis (GSEA) and the Gene Ontology (GO) were performed to the selected genes. Moreover, survival analysis was used to estimate uveal melanoma dataset.
Results
In total, 509 DEGs were identified in GSE24673 (retinoblastoma), 305 DEGs were identified in GSE44295 (uveal melanoma), and 753 DEGs were identified in GSE103439 (basal cell carcinoma of the eyelid). Among those genes, only IGF2BP3 was shared for the three cancer types. A total of 20 DEGs were identified through gene integration (score < 0.05) and IGF2BP3 was ranked the top. Moreover, GO analysis results showed that the 20 DEGs were significantly enriched in WNT signaling pathway, DNA damage, and apoptotic process. GSEA showed that pathways related with cellular respiratory chain are differentially enriched in IGF2BP3 low expression phenotype. Finally, two genes (ID3 and SLC6A15) can predict the overall survival in uveal melanoma patients.
Conclusions
This findings and results of study showed that the identification of DEGs and key pathways gives a promotion to understand the molecular mechanisms underlying the development of ocular cancers, which contribute to a more comprehensive understanding of cancers of the eye and provide new insights for these studies at gene level.









Similar content being viewed by others
Abbreviations
- GEO:
-
Gene Expression Omniniub
- DEGs:
-
differentially expressed gene
- GSEA:
-
gene set enrichment analysis
- GO:
-
Gene Ontology
- CC:
-
cellular component
- BP:
-
biological processes
- MF:
-
molecular function
- TCGA-UVM:
-
The Cancer Genome Atlas-uveal melanomas
References
Parkin DM WS, Ferlay J, Teppo L. 2002. Cancer incidence in five continents. Volume VIII. IARC scientific publications:1–781
Singh AD, Turell ME, Topham AK (2011) Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology 118:1881–1885
Stang A, Parkin DM, Ferlay J, Jockel KH (2005) International uveal melanoma incidence trends in view of a decreasing proportion of morphological verification. Int J Cancer 114:114–123
Bol KF, Mensink HW, Aarntzen EH, Schreibelt G, Keunen JE, Coulie PG, de Klein A, Punt CJ, Paridaens D, Figdor CG, de Vries IJ (2014) Long overall survival after dendritic cell vaccination in metastatic uveal melanoma patients. Am J Ophthalmol 158:939–947
Rietschel P, Panageas KS, Hanlon C, Patel A, Abramson DH, Chapman PB (2005) Variates of survival in metastatic uveal melanoma. J Clin Oncol Off J Am Soc Clin Oncol 23:8076–8080
Ganguly A, Shields CL (2010) Differential gene expression profile of retinoblastoma compared to normal retina. Mol Vis 16:1292–1303
Wang QL, Chen X, Zhang MH, Shen QH, Qin ZM (2015) Identification of hub genes and pathways associated with retinoblastoma based on co-expression network analysis. Genet Mol Res 14:16151–16161
Eagle RC Jr (2013) The pathology of ocular cancer. Eye 27:128–136
Pe’er J (2016) Pathology of eyelid tumors. Indian J Ophthalmol 64:177–190
Milman T, McCormick SA (2013) The molecular genetics of eyelid tumors: recent advances and future directions. Graefes Arch Clin Exp Ophthalmol:419–433
Wang A, Zhang G (2017) Differential gene expression analysis in glioblastoma cells and normal human brain cells based on GEO database. Oncol Lett 14:6040–6044
Kolde R, Laur S, Adler P, Vilo J (2012) Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28:573–580
Wu H, Zhang J (2018) Decreased expression of TFAP2B in endometrial cancer predicts poor prognosis: a study based on TCGA data. Gynecol Oncol 149:592–597
Shen G, He P, Mao Y, Li P, Luh F, Ding G, Liu X, Yen Y (2017) Overexpression of uridine-cytidine kinase 2 correlates with breast cancer progression and poor prognosis. J Breast Cancer 20:132–141
Danda R, Ganapathy K, Sathe G, Madugundu AK, Krishnan UM, Khetan V, Rishi P, Gowda H, Pandey A, Subramanian K, Prasad TK, Elchuri SV (2018) Membrane proteome of invasive retinoblastoma: differential proteins and biomarkers proteomics clinical applications:e1700101
Prabhakaran VC, Gupta A, Huilgol SC, Selva D (2007) Basal cell carcinoma of the eyelids. Compr Ophthalmol Updat 8:1–14
Iwasaki JK, Srivastava D, Moy RL, Lin HJ, Kouba DJ (2012) The molecular genetics underlying basal cell carcinoma pathogenesis and links to targeted therapeutics. J Am Acad Dermatol 66:e167–e178
Soddu S, Di Felice E, Cabras S, Castellanos ME, Atzori L, Faa G, Pilloni L (2013) IMP-3 expression in keratoacanthomas and squamous cell carcinomas of the skin: an immunohistochemical study. Eur J Histochem 57:e6
Lederer M, Bley N, Schleifer C, Huttelmaier S (2014) The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol 29:3–12
Yuhang Zhou, Tingting Huang, Ho Lam Siu, Chi Chun Wong, Yujuan Dong, Feng Wu, Bin Zhang, William K. K. Wu, Alfred S. L. Cheng, Jun Yu, Ka Fai To, Wei Kang (2017) IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer:77
Pryor JG, Simon RA, Bourne PA, Spaulding BO, Scott GA, Xu H (2009) Merkel cell carcinoma expresses K homology domain-containing protein overexpressed in cancer similar to other high-grade neuroendocrine carcinomas. Hum Pathol 40:238–243
Meyer T (2009) Molecular events in skin cancer. Cancer Treat Res 146:189–192
Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, Granter SR, Ramaswamy S, Golub T, Duncan LM, Wagner SN, Brennan C, Chin L (2010) Integrative genome comparison of primary and metastatic melanomas. PLoS One 5:e10770
Cao J, Mu Q, Huang H (2018) The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int 2018:4217259
Oursler MJ, Westendorf JJ, Weivoda MM, Ruan M, Hachfeld CM, Howe A, Davey R, Zajac J, Williams BO, Khosla S (2015) Response to Wnt signaling pathways. J Bone Miner Res Off J Am Soc Bone Miner Res 30:2135–2136
Duchartre Y, Kim YM, Kahn M (2016) The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol 99:141–149
Norbury CJ, Zhivotovsky B (2004) DNA damage-induced apoptosis. Oncogene 23:2797–2808
Tang HL, Tang HM, Mak KH, Hu S, Wang SS, Wong KM, Wong CS, Wu HY, Law HT, Liu K, Talbot CC Jr, Lau WK, Montell DJ, Fung MC (2012) Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell 23:2240–2252
Roos WP, Kaina B (2006) DNA damage-induced cell death by apoptosis. Trends Mol Med 12:440–450
Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5:603–614
Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, Todorova-Manova K, Gerald WL, Brogi E, Benezra R, Massague J (2007) ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A 104:19506–19511
Kamalian L, Forootan SS, Bao ZZ, Zhang Y, Gosney JR, Foster CS, Ke Y (2010) Inhibition of tumourigenicity of small cell lung cancer cells by suppressing Id3 expression. Int J Oncol 37:595–603
Phi JH, Choi SA, Lim SH, Lee J, Wang KC, Park SH, Kim SK (2013) ID3 contributes to cerebrospinal fluid seeding and poor prognosis in medulloblastoma. BMC Cancer 13:291
Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, Min BH, Kim DH, Son HJ, Rhee PL, Kim JJ, Rhee JC, Kim HC, Chun HK, Grady WM, Kim YS (2011) Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol 18:2338–2347
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wan, Q., Tang, J. Exploration of potential key pathways and genes in multiple ocular cancers through bioinformatics analysis. Graefes Arch Clin Exp Ophthalmol 257, 2329–2341 (2019). https://doi.org/10.1007/s00417-019-04410-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-019-04410-2