Abstract
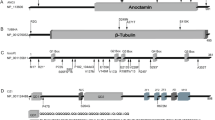
Dynein, cytoplasmic 1, heavy chain 1 (DYNC1H1) encodes a necessary subunit of the cytoplasmic dynein complex, which traffics cargo along microtubules. Dominant DYNC1H1 mutations are implicated in neural diseases, including spinal muscular atrophy with lower extremity dominance (SMA-LED), intellectual disability with neuronal migration defects, malformations of cortical development, and Charcot–Marie–Tooth disease, type 2O. We hypothesized that additional variants could be found in these and novel motoneuron and related diseases. Therefore, we analyzed our database of 1024 whole exome sequencing samples of motoneuron and related diseases for novel single nucleotide variations. We filtered these results for significant variants, which were further screened using segregation analysis in available family members. Analysis revealed six novel, rare, and highly conserved variants. Three of these are likely pathogenic and encompass a broad phenotypic spectrum with distinct disease clusters. Our findings suggest that DYNC1H1 variants can cause not only lower, but also upper motor neuron disease. It thus adds DYNC1H1 to the growing list of spastic paraplegia related genes in microtubule-dependent motor protein pathways.




Similar content being viewed by others
References
Vissers LELM, de Ligt J, Gilissen C et al (2010) A de novo paradigm for mental retardation. Nat Genet 42(12):1109–1112
Willemsen MH, Vissers LEL, Willemsen MAAP et al (2012) Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J Med Genet 49(3):179–183
Weedon MN, Hastings R, Caswell R et al (2011) Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 89(2):308–312
Harms MB, Ori-McKenney KM, Scoto M et al (2012) Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology 78(22):1714–1720
Tsurusaki Y, Saitoh S, Tomizawa K et al (2012) A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics 13(4):327–332
Scoto M, Rossor A, Harms M et al (2015) Novel mutations expand the clinical spectrum of DYNC1H1-associated spinal muscular atrophy. Neurology 84(7):668–679
Poirier K, Lebrun N, Broix L et al (2013) Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet 45(6):639–647
Fiorillo C, Moro F, Yi J et al (2013) Novel dynein DYNC1H1 neck and motor domain mutations link distal SMA and abnormal cortical development. Hum Mutat 35(3):298–302
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595
McKenna A, Matthew H, Banks E et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Gonzalez MA, Acosta Lebrigio RF, Van Booven D et al (2013) GEnomes Management Application (GEM.app): a new software tool for large-scale collaborative genome analysis. Hum Mutat 34(6):842–846
Auer-Grumbach M, Löscher WN, Wagner K et al (2000) Phenotypic and genotypic heterogeneity in hereditary motor neuronopathy type V: a clinical, electrophysiological and genetic study. Brain 123:1612–1623
Copeliovitch L, Katz K, Arbel N, Harries N, Bar-On E, Soudry M (2001) Musculoskeletal deformities in Behr syndrome. J Pediatr Orthop 21:512–514
Stevanin G, Azzedine H, Denora P et al (2008) Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain 131(3):772–784
Schüle R, Schlipf N, Synofzik M et al (2009) Frequency and phenotype of SPG11 and SPG15 in complicated hereditary spastic paraplegia. J Neurol Neurosurg Psychiatry 80(12):1402–1404
Rodefeld MD, Soper NJ (1998) Parahiatal hernia with volvulus and incarceration: laparoscopic repair of a rare defect. J Gastrointest Surg 2(2):193–197
Oates EC, Reddel S, Rodriguez ML et al (2012) Autosomal dominant congenital spinal muscular atrophy: a true form of spinal muscular atrophy caused by early loss of anterior horn cells. Brain 135(Pt 6):1714–1723
Mercuri E, Messina S, Kinali M et al (2004) Congenital form of spinal muscular atrophy predominantly affecting the lower limbs: a clinical and muscle MRI study. Neuromuscul Disord 14:125–129
Synofzik M, Martinez-Carrera LA, Lindig T, Schöls L, Wirth B (2013) Dominant spinal muscular atrophy due to BICD2: a novel mutation refines the phenotype. J Neurol Neurosurg Psychiatry 85(5):590–592
Oates EC, Rossor AM, Havezparast M et al (2013) Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am J Hum Genet 92(6):965–973
Eskuri JM, Stanley CM, Moore SA, Mathews KD (2012) Infantile onset CMT2D/dSMA V in monozygotic twins due to a mutation in the anticodon-binding domain of GARS. J Peripher Nerv Syst 17:132–134
Kanning KC, Kaplan A, Henderson CE (2010) Motor neuron diversity in development and disease. Annu Rev Neurosci 33:409–440
Ciciliot S, Rossi AC, Dyar KA, Blaauw B (2013) Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol 45:2192–2199
Brockington A, Ning K, Heath PR et al (2013) Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol 125:95–109
Blackstone C, O’Kane CJ, Reid E (2011) Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci 12(1):31–42
Timmerman V, Clowes VE, Reid E (2013) Overlapping molecular pathological themes link Charcot-Marie-Tooth neuropathies and hereditary spastic paraplegias. Exp Neurol 246:14–25
Sivagurunathan S, Schnittker RR, Razafsky DS, Nandini S, Plamann MD, King SJ (2012) Analysis of dynein heavy chain mutations reveal complex interactions between dynein motor domains and cellular dynein functions. Genetics 191:1157–1179
Ilieva HS, Yamanaka K, Malkmus S et al (2008) Mutant dynein (Loa) triggers proprioceptive axon loss that extends survival only in the SOD1 ALS model with highest motor neuron death. Proc Natl Acad Sci 105(34):12599–12604
Ori-McKenney KM, Valle RB (2011) Neuronal migration defects in the Loa dynein mutant mouse. Neural Dev 6:26
Deng W, Garret C, Dombert B et al (2010) Neurodegenerative mutation in cytoplasmic dynein alters its organization and dynein-dynactin and dynein-kinesin interactions. J Biol Chem 285(51):39922–39934
Hafezparast M, Klocke R, Ruhrberg C et al (2003) Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science 300(5620):808–812
Chen XJ, Levedakou EN, Millen KJ, Wollmann RL, Soliven B, Popko B (2007) Proprioceptive sensory neuropathy in mice with a mutation in the cytoplasmic Dynein heavy chain 1 gene. J Neurosci 27(52):14515–14524
Insinna C, Baye LM, Amsterdam A, Besharse JC, Link BA (2010) Analysis of a zebrafish dync1h1 mutant reveals multiple functions for cytoplasmic dynein 1 during retinal photoreceptor development. Neural Dev 5:12
Langworthy MM, Appel B (2012) Schwann cell myelination requires Dynein function. Neural Dev 7:37
Acknowledgments
We are grateful for the participation of the patients and families in this study. This work was supported by the Austrian Science Fund (FWF, P23223-B19). M.H. is supported by the National Institute of Neurological Disorders and Stroke/National Institute of Health Grant K08-NS075094. Additional contributions come from the Interdisciplinary Center for Clinical Research, Tübingen (Grant 2191-0-0 to M.S. and Grant 1970-0-0 to R.S.), and the European Union (PIOF-GA-2012- 326681 HSP/CMT genetics and E-Rare-Network NEUROLIPID 01GM1408B to R.S.). S.Z. and A.V.S. are supported by National Institute of Health Grants U54NS0657, R01NS075764, R01NS072248, the Muscular Dystrophy Association, and the CMT Association. M.E.S. is supported by National Institute of Health Grants U54NS0657, R01NS075764, the MDA, and the CMT Association. S.A.M is supported in part by National Institute of Health Grant U54NS053672.
Conflicts of interest
These authors declare that they have no conflict of interest.
Ethical standards
These studies have been approved by the appropriate local ethics committees and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Strickland, A.V., Schabhüttl, M., Offenbacher, H. et al. Mutation screen reveals novel variants and expands the phenotypes associated with DYNC1H1 . J Neurol 262, 2124–2134 (2015). https://doi.org/10.1007/s00415-015-7727-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-015-7727-2