Abstract
Background
Sudden cardiac death (SCD) is a major public health problem and constitutes a diagnostic and preventive challenge in forensic pathology, especially for cases with structural normal hearts at autopsy, so-called sudden arrhythmic death syndrome (SADS). The identification of new genetic risk factors that predispose to SADS is important, because they may contribute to establish the diagnosis and increase the understanding of disease pathways underlying SADS. Pathogenic mutations in the protein coding regions of cardiac genes were found in relation to SADS. However, much remains unknown about variants in non-coding regions of the genome.
Methods and results
In this study, we explored the potential of whole genome sequencing (WGS) and whole transcriptome sequencing (WTS) to find DNA variants in SCD victims with structural normal hearts.
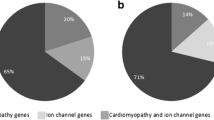
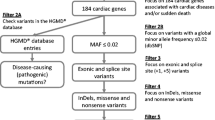
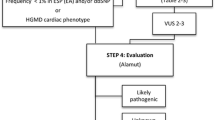
With focus on the non-coding regulatory regions, we re-examined a cohort of 13 SADS and sudden unexplained death in infancy (SUDI) victims without disease causing DNA variants in recognized cardiac genes. The genetic re-examination of DNA was carried out using frozen tissue samples and WTS was carried out using five distinct formalin fixed and paraffin embedded (FFPE) cardiac tissue samples from each individual, including anterior and posterior walls of the left ventricle, ventricular papillary muscle, septum, and the right ventricle. We identified 23 candidate variants in regulatory sequences of cardiac genes, including a variant in the promotor region of NEXN, c.-194A>G, that was found to be statistically significantly (p < 0.05) associated with decreased expression of NEXN and cardiac hypertrophy.
Conclusion
With the use of post-mortem FFPE tissues, we highlight the potential of using WTS investigations and compare gene expression levels with DNA variation in regulatory non-coding regions of the genome for a better understanding of the genetics of cardiac diseases leading to SCD.


Similar content being viewed by others
Abbreviations
- ACMG:
-
American College of Medical Genetics and Genomics
- AWLV:
-
Anterior walls of the left ventricle
- BrS:
-
Brugada syndrome
- CPVT:
-
Catecholaminergic polymorphic ventricular tachycardia
- ENCODE:
-
Encyclopedia of DNA Elements
- DCM:
-
Dilated cardiomyopathy
- FFPE:
-
Formalin fixed and paraffin embedded
- GTEx:
-
The Genotype-Tissue Expression Project
- GTF:
-
Gene transfer format
- HCM:
-
Hypertrophic cardiomyopathy
- LQTS:
-
Long QT syndrome
- Mt.:
-
Mitochondrial
- NUMT:
-
Nuclear mitochondrial DNA segment
- PCA:
-
Principal component analysis
- PM:
-
Papillary muscle
- PMI:
-
Post-mortem interval
- PWLV:
-
Posterior walls of the left ventricle
- SADS:
-
Sudden arrhythmic death syndrome
- SCD:
-
Sudden cardiac death
- SD:
-
Sudden death
- SEP:
-
Septum
- SIDS:
-
Sudden infant death syndrome
- SUDEP:
-
Sudden unexplained death in epilepsy
- SUDI:
-
Sudden unexplained death in infancy
- TFBS:
-
Transcription factor binding sites
- TPM:
-
Transcripts per kilo base million
- VUS:
-
Variants of unknown significance
- WGS:
-
Whole genome sequencing
- WTS:
-
Whole transcriptome sequencing
References
Campuzano O, Allegue C, Partemi S, Iglesias A, Oliva A, Brugada R (2014) Negative autopsy and sudden cardiac death. Int J Legal Med 128:599–606. https://doi.org/10.1007/s00414-014-0966-4
Bjune T, Risgaard B, Kruckow L, Glinge C, Ingemann-Hansen O, Leth PM, Linnet K, Banner J, Winkel BG, Tfelt-Hansen J (2018) Post-mortem toxicology in young sudden cardiac death victims: a nationwide cohort study. Europace 20:614–621. https://doi.org/10.1093/europace/euw435
Semsarian C, Ingles J, Wilde AA (2015) Sudden cardiac death in the young: the molecular autopsy and a practical approach to surviving relatives. Eur Heart J 36:1290–1296. https://doi.org/10.1093/eurheartj/ehv063
Bagnall RD, Weintraub RG, Ingles J, Duflou J, Yeates L, Lam L, Davis AM, Thompson T, Connell V, Wallace J, Naylor C, Crawford J, Love DR, Hallam L, White J, Lawrence C, Lynch M, Morgan N, James P, du Sart D, Puranik R, Langlois N, Vohra J, Winship I, Atherton J, McGaughran J, Skinner JR, Semsarian C (2016) A prospective study of sudden cardiac death among children and young adults. N Engl J Med 374:2441–2452. https://doi.org/10.1056/NEJMoa1510687
Basso C, Aguilera B, Banner J et al (2017) Guidelines for autopsy investigation of sudden cardiac death: 2017 update from the Association for European Cardiovascular Pathology. Virchows Arch 471:691–705. https://doi.org/10.1007/s00428-017-2221-0
Lahrouchi N, Raju H, Lodder EM, Papatheodorou E, Ware JS, Papadakis M, Tadros R, Cole D, Skinner JR, Crawford J, Love DR, Pua CJ, Soh BY, Bhalshankar JD, Govind R, Tfelt-Hansen J, Winkel BG, van der Werf C, Wijeyeratne YD, Mellor G, Till J, Cohen MC, Tome-Esteban M, Sharma S, Wilde AAM, Cook SA, Bezzina CR, Sheppard MN, Behr ER (2017) Utility of post-mortem genetic testing in cases of sudden arrhythmic death syndrome. J Am Coll Cardiol 69:2134–2145. https://doi.org/10.1016/j.jacc.2017.02.046
Bezzina CR, Lahrouchi N, Priori SG (2015) Genetics of sudden cardiac death. Circ Res 116:1919–1936. https://doi.org/10.1161/CIRCRESAHA.116.304030
Tester DJ, Wong LCH, Chanana P, Jaye A, Evans JM, FitzPatrick DR, Evans MJ, Fleming P, Jeffrey I, Cohen MC, Tfelt-Hansen J, Simpson MA, Behr ER, Ackerman MJ (2018) Cardiac genetic predisposition in sudden infant death syndrome. J Am Coll Cardiol 71:1217–1227. https://doi.org/10.1016/j.jacc.2018.01.030
Christiansen SL, Hertz CL, Ferrero-Miliani L, Dahl M, Weeke PE, LuCamp, Ottesen GL, Frank-Hansen R, Bundgaard H, Morling N (2016) Genetic investigation of 100 heart genes in sudden unexplained death victims in a forensic setting. Eur J Hum Genet 24:1797–1802. https://doi.org/10.1038/ejhg.2016.118
Neubauer J, Lecca MR, Russo G, Bartsch C, Medeiros-Domingo A, Berger W, Haas C (2017) Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur J Hum Genet 25:404–409. https://doi.org/10.1038/ejhg.2016.199
Neubauer J, Lecca MR, Russo G, Bartsch C, Medeiros-Domingo A, Berger W, Haas C (2018) Exome analysis in 34 sudden unexplained death (SUD) victims mainly identified variants in channelopathy-associated genes. Int J Legal Med 132:1057–1065. https://doi.org/10.1007/s00414-018-1775-y
van den Boogaard M, Wong LY, Tessadori F et al (2012) Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest 122:2519–2530. https://doi.org/10.1172/JCI62613
Yagihara N, Watanabe H, Barnett P, Duboscq-Bidot L, Thomas AC, Yang P, Ohno S, Hasegawa K, Kuwano R, Chatel S, Redon R, Schott JJ, Probst V, Koopmann TT, Bezzina CR, Wilde AAM, Nakano Y, Aiba T, Miyamoto Y, Kamakura S, Darbar D, Donahue BS, Shigemizu D, Tanaka T, Tsunoda T, Suda M, Sato A, Minamino T, Endo N, Shimizu W, Horie M, Roden DM, Makita N (2016) Variants in the SCN5A promoter associated with various arrhythmia phenotypes. J Am Heart Assoc 5. https://doi.org/10.1161/JAHA.116.003644
Wang J, Wang F, Zhu J, Song M, An J, Li W (2018) Transcriptome profiling reveals PHLDA1 as a novel molecular marker for ischemic cardiomyopathy. J Mol Neurosci 65:102–109. https://doi.org/10.1007/s12031-018-1066-6
Hedegaard J, Thorsen K, Lund MK, Hein AMK, Hamilton-Dutoit SJ, Vang S, Nordentoft I, Birkenkamp-Demtröder K, Kruhøffer M, Hager H, Knudsen B, Andersen CL, Sørensen KD, Pedersen JS, Ørntoft TF, Dyrskjøt L (2014) Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 9:e98187. https://doi.org/10.1371/journal.pone.0098187
Esteve-Codina A, Arpi O, Martinez-Garcia M et al (2017) A comparison of RNA-Seq results from paired formalin-fixed paraffin-embedded and fresh-frozen glioblastoma tissue samples. PLoS One 12:e0170632. https://doi.org/10.1371/journal.pone.0170632
Ferreira PG, Munoz-Aguirre M, Reverter F et al (2018) The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat Commun 9:490. https://doi.org/10.1038/s41467-017-02772-x
Hertz CL, Christiansen SL, Larsen MK, Dahl M, Ferrero-Miliani L, Weeke PE, Pedersen O, Hansen T, Grarup N, Ottesen GL, Frank-Hansen R, Banner J, Morling N (2016) Genetic investigations of sudden unexpected deaths in infancy using next-generation sequencing of 100 genes associated with cardiac diseases. Eur J Hum Genet 24:817–822. https://doi.org/10.1038/ejhg.2015.198
Lindgreen S (2012) AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 5:337. https://doi.org/10.1186/1756-0500-5-337
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv 1303.3997
Van der Auwera GA, Carneiro MO, Hartl C et al (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1-33. https://doi.org/10.1002/0471250953.bi1110s43
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Ienasescu H, Li K, Andersson R, Vitezic M, Rennie S, Chen Y, Vitting-Seerup K, Lagoni E, Boyd M, Bornholdt J, de Hoon MJL, Kawaji H, Lassmann T, The FANTOM Consortium, Hayashizaki Y, Forrest ARR, Carninci P, Sandelin A (2016) On-the-fly selection of cell-specific enhancers, genes, miRNAs and proteins across the human body using SlideBase. Database (Oxford) 2016. https://doi.org/10.1093/database/baw144
Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P (2018) Ensembl 2018. Nucleic Acids Res 46:D754–DD61. https://doi.org/10.1093/nar/gkx1098
Fantom C, the RP, Clst et al (2014) A promoter-level mammalian expression atlas. Nature 507:462–470. https://doi.org/10.1038/nature13182
Encode Project C (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. https://doi.org/10.1038/nature11247
Roadmap Epigenomics C, Kundaje A, Meuleman W et al (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. https://doi.org/10.1038/nature14248
Visel A, Minovitsky S, Dubchak I, Pennacchio LA (2007) VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35:D88–D92. https://doi.org/10.1093/nar/gkl822
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334
Farre D, Roset R, Huerta M et al (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31:3651–3653
Wingender E, Dietze P, Karas H, Knuppel R (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24:238–241
Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858. https://doi.org/10.1038/nature07730
Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394. https://doi.org/10.1089/1066527041410418
Bhattacharya A, Ziebarth JD, Cui Y (2014) PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 42:D86–D91. https://doi.org/10.1093/nar/gkt1028
Layer RM, Chiang C, Quinlan AR, Hall IM (2014) LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:R84. https://doi.org/10.1186/gb-2014-15-6-r84
Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, Marth GT, Quinlan AR, Hall IM (2015) SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods 12:966–968. https://doi.org/10.1038/nmeth.3505
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774. https://doi.org/10.1101/gr.135350.111
Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010) RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26:493–500. https://doi.org/10.1093/bioinformatics/btp692
Hertz CL, Christiansen SL, Ferrero-Miliani L et al (2016) Next-generation sequencing of 100 candidate genes in young victims of suspected sudden cardiac death with structural abnormalities of the heart. Int J Legal Med 130:91–102. https://doi.org/10.1007/s00414-015-1261-8
Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, Liu X, Veinot JP, Tang ASL, Stewart AFR, Tesson F, Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L, Bai D (2006) Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med 354:2677–2688. https://doi.org/10.1056/NEJMoa052800
Olesen MS, Andreasen L, Jabbari J, Refsgaard L, Haunsø S, Olesen SP, Nielsen JB, Schmitt N, Svendsen JH (2014) Very early-onset lone atrial fibrillation patients have a high prevalence of rare variants in genes previously associated with atrial fibrillation. Heart Rhythm 11:246–251. https://doi.org/10.1016/j.hrthm.2013.10.034
Lubkemeier I, Andrie R, Lickfett L et al (2013) The Connexin40A96S mutation from a patient with atrial fibrillation causes decreased atrial conduction velocities and sustained episodes of induced atrial fibrillation in mice. J Mol Cell Cardiol 65:19–32. https://doi.org/10.1016/j.yjmcc.2013.09.008
Kanthan A, Fahmy P, Rao R, Pouliopoulos J, Alexander IE, Thomas SP, Kizana E (2018) Human Connexin40 mutations slow conduction and increase propensity for atrial fibrillation. Heart Lung Circ 27:114–121. https://doi.org/10.1016/j.hlc.2017.02.010
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980–D985. https://doi.org/10.1093/nar/gkt1113
Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, Grimmler M, Liptau H, Hetzer R, Regitz-Zagrosek V, Fischer C, Nürnberg P, Schunkert H, Katus HA, Rottbauer W (2009) Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med 15:1281–1288. https://doi.org/10.1038/nm.2037
Wang H, Li Z, Wang J, Sun K, Cui Q, Song L, Zou Y, Wang X, Liu X, Hui R, Fan Y (2010) Mutations in NEXN, a Z-disc gene, are associated with hypertrophic cardiomyopathy. Am J Hum Genet 87:687–693. https://doi.org/10.1016/j.ajhg.2010.10.002
Jansen JA, van Veen TA, de Bakker JM, van Rijen HV (2010) Cardiac connexins and impulse propagation. J Mol Cell Cardiol 48:76–82. https://doi.org/10.1016/j.yjmcc.2009.08.018
Hew KW, Keller KA (2003) Postnatal anatomical and functional development of the heart: a species comparison. Birth Defects Res B Dev Reprod Toxicol 68:309–320. https://doi.org/10.1002/bdrb.10034
Acknowledgements
We thank Carina Grøntved Jønck for bioinformatics support, Anja Ladegaard Jørgensen for technical assistance in the laboratory, Mikkel M. Andersen for statistical support, and Steffan N. Christiansen and Sofie L. Christiansen for fruitful discussions. This work was supported by Ellen and Aage Andersen’s Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics
All procedures performed in the study were in accordance with the ethical standards of the Committees on Health Research Ethics in the Capital Region of Denmark (H-2-2012-017) and the Danish Data Protection Agency (2011-54-1262).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
Boxplot of the percentage of mitochondrial (mt)-RNA in cardiac tissue from four sudden unexplained death in infancy (SUDI) and six sudden arrhythmic death syndrome (SADS) victims. mt-RNA was identified using the gene transfer format file of the hg19 reference genome from the GENCODE consortium release 27. The whiskers extends 1.5 * IQR from the hinge (where IQR is the inter-quartile range, or distance between the first and third quartiles). (JPEG 40 kb)
Supplementary Figure 2
Principle component analysis of all protein coding transcripts in four sudden unexplained death in infancy (SUDI) and six sudden arrhythmic death syndrome (SADS) victims. Principle components represents variation between gene expression levels of SUDI and SADS victims. (JPEG 116 kb)
Supplementary Figure 3
Boxplot of the percentage of mitochondrial (mt)-RNA in five mildly decomposed sudden cardiac death victims and five sudden cardiac death victims characterized as not decomposed. Decomposition status was determined by a forensic pathologist at autopsy. mt-RNA was identified using the gene transfer format file of the hg19 reference genome from the GENCODE consortium release 27. The whiskers extends 1.5 * IQR from the hinge (where IQR is the inter-quartile range, or distance between the first and third quartiles). (JPEG 48 kb)
Supplementary Table 1
Detailed description of cases in the study population. (XLSX 18 kb)
Supplementary Table 2
Nuclear mitochondrial DNA segment (NUMT) genes excluded from analysis. Positions are from the human genome assembly GRCh37 (hg19). (XLSX 68 kb)
Supplementary Table 3
Whole transcriptome sequencing and alignment statistics of the anterior and posterior wall of the left ventricle, right ventricle, septum, and ventricular papillary muscle of 10 sudden cardiac death victims. Alignment statistics were obtained from STAR (Spliced Transcripts Alignment to a Reference), and RNA types were extracted from the gene transfer format file of the hg19 reference genome from the GENCODE consortium release 27. Abbreviations: Mb = Megabase, PF = Passing Filter, RIN = RNA Integrity Number. (XLSX 51 kb)
Supplementary Table 4
(DOCX 30 kb)
Rights and permissions
About this article
Cite this article
Andersen, J.D., Jacobsen, S.B., Trudsø, L.C. et al. Whole genome and transcriptome sequencing of post-mortem cardiac tissues from sudden cardiac death victims identifies a gene regulatory variant in NEXN. Int J Legal Med 133, 1699–1709 (2019). https://doi.org/10.1007/s00414-019-02127-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-019-02127-9