Abstract
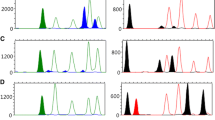
Recent advances in whole-genome epigenetic analysis indicate that chromosome segments called tissue-specific differentially methylated regions (tDMRs) show different DNA methylation profiles according to cell or tissue type. Therefore, body fluid-specific differential DNA methylation is a promising indicator for body fluid identification. However, DNA methylation patterns are susceptible to change in response to environmental factors and aging. Therefore, we investigated age-related methylation changes in semen-specific tDMRs using body fluids from young and elderly men. After confirming the stability of the body fluid-specific DNA methylation profile over time, two different multiplex PCR systems were constructed using methylation-sensitive restriction enzyme PCR and methylation SNaPshot, in order to analyze the methylation status of specific CpG sites from the USP49, DACT1, PRMT2, and PFN3 tDMRs. Both multiplex systems could successfully identify semen with spermatozoa and could differentiate menstrual blood and vaginal fluids from blood and saliva. Although including more markers for body fluid identification might be necessary, this study adds to the support that body fluid identification by DNA methylation profiles could be a valuable tool for forensic analysis of body fluids.




Similar content being viewed by others
References
Kayser M, de Knijff P (2011) Improving human forensics through advances in genetics, genomics and molecular biology. Nat Rev Genet 12:179–192
Virkler K, Lednev IK (2009) Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int 188:1–17
Bauer M, Polzin S, Patzelt D (2003) Quantification of RNA degradation by semi-quantitative duplex and competitive RT-PCR: a possible indicator of the age of bloodstains? Forensic Sci Int 138:94–103
Bauer M, Patzelt D (2002) Evaluation of mRNA markers for the identification of menstrual blood. J Forensic Sci 47:1278–1282
Juusola J, Ballantyne J (2003) Messenger RNA profiling: a prototype method to supplant conventional methods for body fluid identification. Forensic Sci Int 135:85–96
Juusola J, Ballantyne J (2005) Multiplex mRNA profiling for the identification of body fluids. Forensic Sci Int 152:1–12
Nussbaumer C, Gharehbaghi-Schnell E, Korschineck I (2006) Messenger RNA profiling: a novel method for body fluid identification by real-time PCR. Forensic Sci Int 157:181–186
Juusola J, Ballantyne J (2007) mRNA profiling for body fluid identification by multiplex quantitative RT-PCR. J Forensic Sci 52:1252–1262
Zubakov D, Hanekamp E, Kokshoorn M, van Ijcken W, Kayser M (2008) Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int J Legal Med 122:135–142
Hanson EK, Lubenow H, Ballantyne J (2009) Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem 387:303–314
Setzer M, Juusola J, Ballantyne J (2008) Recovery and stability of RNA in vaginal swabs and blood, semen, and saliva stains. J Forensic Sci 53:296–305
Zubakov D, Kokshoorn M, Kloosterman A, Kayser M (2009) New markers for old stains: stable mRNA markers for blood and saliva identification from up to 16-year-old stains. Int J Legal Med 123:71–74
Haas C, Klesser B, Maake C, Bär W, Kratzer A (2009) mRNA profiling for body fluid identification by reverse transcription endpoint PCR and realtime PCR. Forensic Sci Int Genet 3:80–88
Frumkin D, Wasserstrom A, Budowle B, Davidson A (2011) DNA methylation-based forensic tissue identification. Forensic Sci Int Genet 5:517–524
Lee HY, Park MJ, Choi A, An JH, Yang WI, Shin KJ (2012) Potential forensic application of DNA methylation profiling to body fluid identification. Int J Legal Med 126:55–62
Holliday R, Pugh JE (1975) DNA modification mechanisms and gene activity during development. Science 187:226–232
Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321:209–213
Russo VEA, Martienssen RA, Riggs AD (1996) Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, Plainview
Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS (2009) Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 18:4808–4817
Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA (2005) Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A 102:3336–3341
Ohgane J, Yagi S, Shiota K (2008) Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta 29:S29–35
Illingworth R, Kerr A, Desousa D, Jørgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6:e22
Wilson VL, Smith RA, Ma S, Cutler RG (1987) Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem 262:9948–9951
Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y (2004) Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet 68:196–204
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5:e1000602
Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T (2005) BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21:4067–4068
Rohde C, Zhang Y, Jurkowski TP, Stamerjohanns H, Reinhardt R, Jeltsch A (2008) Bisulfite sequencing Data Presentation and Compilation (BDPC) web server—a useful tool for DNA methylation analysis. Nucleic Acids Res 36:e34
Kaminsky Z, Petronis A (2009) Methylation SNaPshot: a method for the quantification of site-specific DNA methylation levels. Meth Mol Biol 507:241–255
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431
You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9:253
Frumkin D, Wasserstrom A, Davidson A, Grafit A (2010) Authentication of forensic DNA samples. Forensic Sci Int Genet 4:95–103
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0005208).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ja Hyun An and Ajin Choi equally contributed to this work.
Rights and permissions
About this article
Cite this article
An, J.H., Choi, A., Shin, KJ. et al. DNA methylation-specific multiplex assays for body fluid identification. Int J Legal Med 127, 35–43 (2013). https://doi.org/10.1007/s00414-012-0719-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-012-0719-1