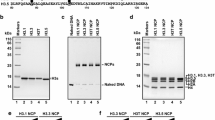
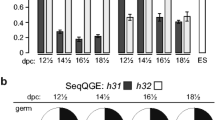
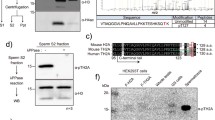
Abstract
The incorporation of histone variants into chromatin plays an important role for the establishment of particular chromatin states. Six human histone H3 variants are known to date, not counting CenH3 variants: H3.1, H3.2, H3.3 and the testis-specific H3.1t as well as the recently described variants H3.X and H3.Y. We report the discovery of H3.5, a novel non-CenH3 histone H3 variant. H3.5 is encoded on human chromosome 12p11.21 and probably evolved in a common ancestor of all recent great apes (Hominidae) as a consequence of H3F3B gene duplication by retrotransposition. H3.5 mRNA is specifically expressed in seminiferous tubules of human testis. Interestingly, H3.5 has two exact copies of ARKST motifs adjacent to lysine-9 or lysine-27, and lysine-79 is replaced by asparagine. In the Hek293 cell line, ectopically expressed H3.5 is assembled into chromatin and targeted by PTM. H3.5 preferentially colocalizes with euchromatin, and it is associated with actively transcribed genes and can replace an essential function of RNAi-depleted H3.3 in cell growth.





Similar content being viewed by others
References
Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20:91–100
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17:1870–1881
Hake SB, Allis CD (2006) Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci USA 103:6428–6435
Henikoff S, Ahmad K (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21:133–153
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Loyola A, Almouzni G (2007) Marking histone H3 variants: how, when and why? Trends Biochem Sci 32:425–433
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–260
Olins DE, Olins AL (2003) Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol 4:809–814
Park YJ, Luger K (2008) Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol 18:282–289
Postberg J, Forcob S, Chang WJ, Lipps HJ (2010) The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol Biol 10:259
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Stehle IM, Postberg J, Rupprecht S, Cremer T, Jackson DA, Lipps HJ (2007) Establishment and mitotic stability of an extra-chromosomal mammalian replicon. BMC Cell Biol 8:33
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF et al (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99:16899–16903
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E et al (2010) Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol 190:777–791
CL,Woodcock, RP Ghosh (2010) Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol 2:10.1101/cshperspect.a000596
Zalmout IS, Sanders WJ, Maclatchy LM, Gunnell GF, Al-Mufarreh YA, Ali MA, Nasser AA, Al-Masari AM, Al-Sobhi SA, Nadhra AO et al (2010) New Oligocene primate from Saudi Arabia and the divergence of apes and old world monkeys. Nature 466:360–364
Acknowledgements
Whole chromosome #12 paint was kindly donated by Dr. Marion Cremer, LMU Munich. Bonobo sperm as well as gorilla saliva samples were kindly provided by Dr. Arne Lawrenz, Wuppertal Zoo. Testicular tissue was sampled and kindly provided by Ulrich Gertenbach and Stephan Roth, HELIOS Klinikum Wuppertal, Urology. This work was supported by the HELIOS Research Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Nigg
Electronic supplementary material
Below is the link to the electronic supplementary material.

Supporting Data 1
Alignment of the protein coding sequences of H3 variant genes used in this study. (JPEG 16451 kb)

Supporting Data 2
Alignment of the H3F3C gene with the mRNA sequences of H3.5 as well as H3.3/H3F3B. (JPEG 2514 kb)

Supporting Data 3
Ectopically expressed H3.5 can replace H3.3 to some extent. a Comparison of doubling times in cells co-transfected with H3F3A/H3F3B siRNAs and the GST-H3.5 expression vector and cells transfected with H3F3A/H3F3B siRNAs only at four successive days. b Nascent BrdU-labelled DNA (green) in cells transfected with H3F3A/H3F3B siRNAs. DAPI (blue) was used for DNA counterstaining. c. Nascent DNA (green) in cells co-transfected with H3F3A/H3F3B siRNAs and the GST-H3.5 expression vector. DAPI (blue) was used for DNA counterstaining. (JPEG 476 kb)

Supporting Data 4
Structural simulation of H3.5 within the nucleosome with emphasis on the chaperone recognition domain. Selected residues are labelled and colour-shaded. (JPEG 809 kb)
Rights and permissions
About this article
Cite this article
Schenk, R., Jenke, A., Zilbauer, M. et al. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120, 275–285 (2011). https://doi.org/10.1007/s00412-011-0310-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-011-0310-4