Abstract
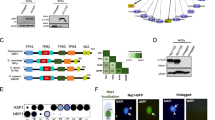
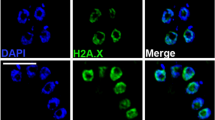
Histone H3 variants play critical roles in the functional specialization of chromatin by epigenetically marking centromeric chromatin and transcriptionally active or silent genes. Specifically, the cenH3 histone variant acts as the primary epigenetic determinant of the site of kinetochore assembly at centromeres. Although the function of histone variants is well studied in plants, animals, and fungi, there is little knowledge of the evolutionary conservation of histone variants and their function in most protists. We find that Giardia intestinalis—a diplomonad parasite with two equivalent nuclei—has two phylogenetically distinct histone H3 variants with N-terminal extensions and nonconserved promoters. To determine their role in chromatin dynamics, conventional H3 and the two H3 variants were GFP-tagged, and their subcellular location was monitored during interphase and mitosis. We demonstrate that one cenH3-like variant has a conserved function in epigenetically marking centromeres. The other H3 variant (H3B) has a punctate distribution on chromosomes, but does not colocalize with active transcriptional regions as indicated by H3K4 methylation. We suggest that H3B could instead mark noncentromeric heterochromatin. Giardia is a member of the Diplomonads and represents an ancient divergence from metazoans and fungi. We confirm the ancient role of histone H3 variants in modulating chromatin architecture, and suggest that monocentric chromosomes represent an ancestral chromosome morphology.





Similar content being viewed by others
References
Adam RD (2000) The Giardia lamblia genome. Int J Parasitol 30(4):475–484
Adam RD (2001) Biology of Giardia lamblia. Clin Microbiol Rev 14(3):447–475
Adam R, Nash T et al (1988) The Giardia lamblia trophozoite contains sets of closely related chromosomes. Nucleic Acids Res 16(10):4555–4567
Ahmad K, Henikoff S (2002a) Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci USA 99(Suppl 4):16477–16484
Ahmad K, Henikoff S (2002b) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9(6):1191–1200
Aiyar A (2000) The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol Biol 132:221–241
Baldauf SL (2003) The deep roots of eukaryotes. Science 300(5626):1703–1706
Baldauf SL, Roger AJ et al (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290(5493):972–977
Bernard P, Maure JF et al (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294(5551):2539–2542
Best AA, Morrison HG et al (2004) Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia. Genome Res 14(8):1537–1547
Brown DT (2001) Histone variants: are they functionally heterogeneous? Genome Biol 2(7):REVIEWS0006
Ciccarelli FD, Doerks T et al (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311(5765):1283–1287
Dacks JB, Marinets A et al (2002) Analyses of RNA Polymerase II genes from free-living protists: phylogeny, long branch attraction, and the eukaryotic big bang. Mol Biol Evol 19(6):830–840
Dernburg AF (2001) Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol 153(6):F33–F38
Elmendorf HG, Dawson SC et al (2003) The cytoskeleton of Giardia lamblia. Int J Parasitol 33(1):3–28
Graczyk TK (2005) Is Giardia a living fossil? Trends Parasitol 21(3):104–107
Henikoff S, Ahmad K (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21:133–153
Kabnick KS, Peattie DA (1990) In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J Cell Sci 95(Pt 3):353–360
Keister DB (1983) Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg 77:487–488
Knight J (2004) Giardia: not so special, after all? Nature 429(6989):236–237
Lo AW, Magliano DJ et al (2001) A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res 11(3):448–457
Maddox PS, Oegema K et al (2004) “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res 12(6):641–653
Malik HS, Henikoff S (2001) Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157(3):1293–1298
Malik HS, Henikoff S (2002) Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev 12(6):711–718
Malik HS, Henikoff S (2003) Phylogenomics of the nucleosome. Nat Struct Biol 10(11):882–891
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6(11):838–849
McKittrick E, Gafken PR et al (2004) Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA 101(6):1525–1530
Pascreau G, Arlot-Bonnemains Y et al (2003) Phosphorylation of histone and histone-like proteins by aurora kinases during mitosis. Prog Cell Cycle Res 5:369–374
Pray-Grant MG, Daniel JA et al (2005) Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433(7024):434–438
Prigent C, Dimitrov S (2003) Phosphorylation of serine 10 in histone H3, what for? J Cell Sci 116(Pt 18):3677–3685
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Sagolla MS, Dawson SC, Mancuso JJ, Cande WZ (2006) Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J Cell Sci 119 (23):4889–4900
Savioli L, Smith H et al (2006) Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative’. Trends Parasitol 22(5):203–208
Singer SM, Yee J et al (1998) Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol 92(1):59–69
Smith MM (2002) Centromeres and histone variant: what, where, when and why? Curr Opin Cell Biol 14(3):279–285
Sogin ML, Gunderson JH et al (1989) Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 243(4887):75–77
Solari AJ, Rahn MI et al (2003) A unique mechanism of nuclear division in Giardia lamblia involves components of the ventral disk and the nuclear envelope. Biocell 27(3):329–346
Sullivan BA, Blower MD et al (2001) Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet 2(8):584–596
Van de Peer Y, Baldauf SL et al (2000) An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J Mol Evol 51(6):565–576
Van Hooser AA, Ouspenski II et al (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114(Pt 19):3529–3542
Wu G, McArthur AG et al (2000) Core histones of the amitochondriate protist, Giardia lamblia. Mol Biol Evol 17(8):1156–1163
Yu LZ, Birky CW Jr et al (2002) The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot Cell 1(2):191–199
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15(18):2343–2360
Acknowledgements
We would like to acknowledge Heidi Elmendorf (Georgetown University), C.C. Wang and colleagues (UCSF), and Keith Gull (Oxford University, UK) for plasmids, reagents, and methodologies. Additionally, we thank Hillary Morrison and other members of the Giardia Genome Project for access to genomic data. We also thank Elizabeth Slawson and members of the Cande lab for the helpful discussions. This work was supported by an NIH grant A1054693 and grant from the University of California Cancer Research Coordinating Committee (CRCC) to WZC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Choo
Appendix
Below is the link to the electronic supplementary material.
Fig. S1
cenH3::GFP foci are visible in 3D stack. This movie steps through individual sections in a 3D stack of images taken at 0.2 μm depth using a cenH3::GFP strain. Individual foci are visible (MOV 2.6 MB).
Rights and permissions
About this article
Cite this article
Dawson, S.C., Sagolla, M.S. & Cande, W.Z. The cenH3 histone variant defines centromeres in Giardia intestinalis . Chromosoma 116, 175–184 (2007). https://doi.org/10.1007/s00412-006-0091-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-006-0091-3