Abstract
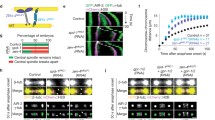
Ndc10p is one of the DNA-binding constituents of the kinetochore in Saccharomyces cerevisiae but light microscopy analysis suggests that Ndc10p is not limited to kinetochore regions. We examined the localization of Ndc10p using immunoelectron microscopy and showed that Ndc10p is associated with spindle microtubules from S-phase through anaphase. By serial section reconstruction of mitotic spindles combined with immunogold detection, we showed that Ndc10p interacts with microtubules laterally as well as terminally. About 50% of the gold label in serial section reconstructions of short mitotic spindles was associated with the walls of spindle microtubules. Interaction of kinetochore components with microtubule walls was also shown for kinetochore protein Ndc80p. Our data suggest that at least a subset of kinetochore-associated protein is dispersed throughout the mitotic spindle.







Similar content being viewed by others
References
Adams IR, Kilmartin JV. (2000) Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol 10:329–335
Andreassen PR, Palmer DK, Wener MH, Margolis RL (1991) Telophase disc: a new mammalian mitotic organelle that bisects telophase cells with a possible function in cytokinesis. J Cell Sci 99:523–534
Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G (2001a) Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol 155:1137–1145
Cheeseman IM, Enquist-Newman M, Müller-Reichert T, Drubin DG, Barnes G (2001b) Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol 152:197–212
Cheeseman IM, Drubin DG, Barnes G (2002) Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol 157:1999–2003
Cooke CA, Heck MMS, Earnshaw WC (1987) The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol 105:2053–2067
Ding R, McDonald KL, McIntosh JR (1993) Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol 120:141–151
Fitzgerald-Hayes M, Clarke L, Carbon J (1982) Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29:235–244
Goh PY, Kilmartin JV (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol 121:503–512
Goshima G, Yanagida M (2000) Establishing biorientation occurs with precocious separation of sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100:619–633
Guacci V, Hogan E, Koshland D (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 125:517–530
He H, Asthana S, Sorger PK (2000) Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101:763–775
He H, Rines DR, Espelin C W, Sorger PK.(2001) Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106:195–206
Hoyt MA, He L, Loo KK, Saunders WS (1992) Two Saccharomyces cerevisiae kinesin-related gene-products required for mitotic spindle assembly. J Cell Biol 118:109–120
Hyman AA, Sorger PK (1995) Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol 11:471–495
Ito H, Fukuda Y, Murat K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168
Janke C, Ortiz J, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, Schramm C, Schiebel E (2001) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 20:777–791
Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E (2002) Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J 21:181–193
Jiang W, Lechner J, Carbon J (1993) Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol 121:513–519
Lechner J, Carbon J (1991) A 240 kD multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64:717–725
Lim HH, Goh PY, Surana U (1998) Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol 8:231–234
McDonald K, Müller-Reichert T (2002) Cryomethods for thin section electron microscopy. Methods Enzymol 351:96–123
McDonald KL, Mastronarde DN, O'Toole ET, Din R, McIntosh JR (1991) Computer-based tools for morphometric analysis of mitotic spindles and other microtubule systems. EMSA Bull 21:47–53
Measday V, Hailey DW, Pot I, Givan S, Hyland KM, Gagney G, Fields S, Davis TN, Hieter P (2002) Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev 16:101–113
Müller-Reichert T, Ashford A, Antony C, Hyman A (2000) Ultrastructural localization of antigens in the mitotic spindle of budding yeast using high pressure freezing and freeze substitution. Microsc Microanal 6 (Suppl 2):300–301
Nasmyth K (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559–565
O'Toole ET, Mastronarde DN, Giddings TH, Winey M, Burke DJ, McIntosh RJ (1997) Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol Biol Cell 8:1–11
Ortiz J, Stemmann O, Rank S, Lechner J (1999) A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev 13:1140–1155
Pankov R, Lemieux M, Hancock R (1990) An antigen located in the kinetochore region in metaphase, and on polar microtubule ends in the midbody region in anaphase, characterized using a monoclonal antibody. Chromosoma 99:95–101
Pereira G, Tanaka TU, Nasmyth K, Schiebel E (2001) Mode of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J 20:6359–6370
Peterson JB, Ris H (1976) Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci 22:219–242
Pringle JR, Adams AEM, Drubin DG, Haarer BK (1991) Immunofluorescence methods for yeast. Methods Enzymol 194:565–602
Quan-wen J, Fuchs J, Loidl J (2000) Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci 113:1903–1912
Rieder CL, Alexander SP (1990) Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol 110:81–95
Roos U-P (1973) Light and electron microscopy of rat kangaroo cells in mitosis: II. Kinetochore structure and function. Chromosoma 41:195–220
Rose MD, Winston F, Hieter P (1990) Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sassoon I, Severin FF Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJ, Sorger PK, Hyman AA (1999) Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev 13:545–555
Schertha H, Loidl J, Schuster T, Schweizer D (1992) Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma 101:590–595
Skibbens RV, Skeen VP, Salmon ED (1993) Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol 122:859–875
Sorger PK, Severin FF, Hyman AA (1994) Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol 127:995–1008
Sorger PK, Doheny KF, Hieter P, Kopski KM, Huffaker TC, Hyman AA (1995) Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci U S A 92:12026–12030
Spang A, Geissler S, Grein K, Schiebel E (1996) γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol 134:429–441
Straight AF, Marshall WF, Sedat JW, Murray AW (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277:574–578
Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K (2002) Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108:317–329
Tavormina PA, Burke DJ (1998) Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics 148:1701–1713
Wigge PA, Kilmartin JV (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152:349–360
Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141:967–977
Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR (1995) Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol 129:1601–1615
Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW(1992) CEN-P is a putative kinetochore motor that accumulates just before mitosis. Nature 359:536–539
Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS (1999) Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol 146:415–425
Acknowledgements
We thank Dr. J. Kilmartin for providing us with the antibody against Ndc80p. We are grateful to Dr. K. McDonald for sharing expertise in high pressure freezing and freeze substitution. We thank Dr. F. Gail (Marine Laboratory, University of Paris 6, Paris, France) for use of the high pressure freezer and J.-P. Lechaire for expert technical assistance. We especially thank Drs. A. Desai, R. Heald, K. McDonald, J.R. McIntosh, J. Paluh, and T. U. Tanaka for their critical comments on the manuscript. Part of this work was supported by the German Research Foundation (DFG grant Mü 1423/1-1 to T. M.-R.) and by the National Institute of Health (grant RR-0592 to J.R. McIntosh). The experiments described in this paper comply with the current laws of the countries in which the experiments were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by: R. Allshire
Rights and permissions
About this article
Cite this article
Müller-Reichert, T., Sassoon, I., O'Toole, E. et al. Analysis of the distribution of the kinetochore protein Ndc10p in Saccharomyces cerevisiae using 3-D modeling of mitotic spindles. Chromosoma 111, 417–428 (2003). https://doi.org/10.1007/s00412-002-0220-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-002-0220-6