Abstract
Background
Asbestos is a carcinogen linked to malignant mesothelioma (MM) and lung cancer. Some gene aberrations related to asbestos exposure are recognized, but many associated mutations remain obscure. We performed exome sequencing to determine the association of previously known mutations (driver gene mutations) with asbestos and to identify novel mutations related to asbestos exposure in lung adenocarcinoma (LAC) and MM.
Methods
Exome sequencing was performed on DNA from 47 tumor tissues of MM (21) and LAC (26) patients, 27 of whom had been asbestos-exposed (18 MM, 9 LAC). In addition, 9 normal lung/blood samples of LAC were sequenced. Novel mutations identified from exome data were validated by amplicon-based deep sequencing. Driver gene mutations in BRAF, EGFR, ERBB2, HRAS, KRAS, MET, NRAS, PIK3CA, STK11, and ephrin receptor genes (EPHA1-8, 10 and EPHB1-4, 6) were studied for both LAC and MM, and in BAP1, CUL1, CDKN2A, and NF2 for MM.
Results
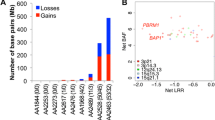
In asbestos-exposed MM patients, previously non-described NF2 frameshift mutation (one) and BAP1 mutations (four) were detected. Exome data mining revealed some genes potentially associated with asbestos exposure, such as MRPL1 and SDK1. BAP1 and COPG1 mutations were seen exclusively in MM. Pathogenic KRAS mutations were common in LAC patients (42 %), both in non-exposed (n = 5) and exposed patients (n = 6). Pathogenic BRAF mutations were found in two LACs.
Conclusion
BAP1 mutations occurred in asbestos-exposed MM. MRPL1, SDK1, SEMA5B, and INPP4A could possibly serve as candidate genes for alterations associated with asbestos exposure. KRAS mutations in LAC were not associated with asbestos exposure.

Similar content being viewed by others
References
IARC (2012) IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Arsenic, metals, fibres and dusts, A review of human carcinogens, p 100C
Kane A, Jean D, Knuutila S et al (2014) Malignant mesothelioma: mechanism of carcinogenesis. In: Anttila S, Boffetta P (eds) Occupational cancers. Springer, London, p 299
Tiainen M, Tammilehto L, Rautonen J et al (1989) Chromosomal abnormalities and their correlations with asbestos exposure and survival in patients with mesothelioma. Br J Cancer 60:618–626
Guled M, Lahti L, Lindholm PM et al (2009) CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer 48:615–623. doi:10.1002/gcc.20669
Kettunen E, Knuutila S (2014) Malignant mesothelioma: molecular markers. In: Anttila S, Boffetta P (eds) Occupational cancers. Springer, London, p 325
Xu J, Kadariya Y, Cheung M et al (2014) Germline mutation of Bap1 accelerates development of asbestos-induced malignant mesothelioma. Cancer Res 74:4388–4397. doi:10.1158/0008-5472.CAN-14-1328
Betti M, Casalone E, Ferrante D et al (2015) Inference on germline BAP1 mutations and asbestos exposure from the analysis of familial and sporadic mesothelioma in a high-risk area. Genes Chromosom Cancer 54:51–62. doi:10.1002/gcc.22218
Nielsen LS, Baelum J, Rasmussen J et al (2014) Occupational asbestos exposure and lung cancer–a systematic review of the literature. Arch Environ Occup Health 69:191–206. doi:10.1080/19338244.2013.863752
Inamura K, Ninomiya H, Nomura K et al (2014) Combined effects of asbestos and cigarette smoke on the development of lung adenocarcinoma: different carcinogens may cause different genomic changes. Oncol Rep 32:475–482. doi:10.3892/or.2014.3263
Husgafvel-Pursiainen K, Hackman P, Ridanpaa M et al (1993) K-ras mutations in human adenocarcinoma of the lung: association with smoking and occupational exposure to asbestos. Int J Cancer 53:250–256
Andujar P, Wang J, Descatha A et al (2010) p16INK4A inactivation mechanisms in non-small-cell lung cancer patients occupationally exposed to asbestos. Lung Cancer 67:23–30. doi:10.1016/j.lungcan.2009.03.018
Wikman H, Ruosaari S, Nymark P et al (2007) Gene expression and copy number profiling suggests the importance of allelic imbalance in 19p in asbestos-associated lung cancer. Oncogene 26:4730–4737 1210270
Nymark P, Guled M, Borze I et al (2011) Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. Genes Chromosom Cancer 50:585–597. doi:10.1002/gcc.20880
Nymark P, Aavikko M, Makila J et al (2013) Accumulation of genomic alterations in 2p16, 9q33.1 and 19p13 in lung tumours of asbestos-exposed patients. Mol Oncol 7:29–40. doi:10.1016/j.molonc.2012.07.006
Karjalainen A, Anttila S, Heikkilä L et al (1993) Asbestos exposure among Finnish lung cancer patients: occupational history and fiber concentration in lung tissue. Am J Ind Med 23:461–471
Tuomi T (1992) Fibrous minerals in the lungs of mesothelioma patients: comparison between data on SEM, TEM, and personal interview information. Am J Ind Med 21:155–162
Tuononen K, Maki-Nevala S, Sarhadi VK et al (2013) Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosom Cancer 52:503–511. doi:10.1002/gcc.22047
Sulonen AM, Ellonen P, Almusa H et al (2011) Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol 12:R94-2011-12-9-r94, DOI: 10.1186/gb-2011-12-9-r94
St John J SeqPrep. https://github.com/jstjohn/SeqPrep
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi:10.1038/nmeth.1923
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and samtools. Bioinformatics 25:2078–2079. doi:10.1093/bioinformatics/btp352
BCFTools. http://samtools.github.io/bcftools/
DePristo MA, Banks E, Poplin R et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi:10.1038/ng.806
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. doi:10.1038/nprot.2009.86
Choi Y, Sims GE, Murphy S et al (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. doi:10.1371/journal.pone.0046688
Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi:10.1093/bib/bbs017
Maki-Nevala S, Kaur Sarhadi V, Tuononen K et al (2013) Mutated ephrin receptor genes in non-small cell lung carcinoma and their occurrence with driver mutations-targeted resequencing study on formalin-fixed, paraffin-embedded tumor material of 81 patients. Genes Chromosom Cancer 52:1141–1149. doi:10.1002/gcc.22109
Bott M, Brevet M, Taylor BS et al (2011) The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 43:668–672. doi:10.1038/ng.855
Zauderer MG, Bott M, McMillan R et al (2013) Clinical characteristics of patients with malignant pleural mesothelioma harboring somatic BAP1 mutations. J Thorac Oncol 8:1430–1433. doi:10.1097/JTO.0b013e31829e7ef9
Hahn Y, Lee YJ, Yun JH et al (2000) Duplication of genes encoding non-clathrin coat protein gamma-COP in vertebrate, insect and plant evolution. FEBS Lett 482:31–36
Sudo H, Tsuji AB, Sugyo A et al (2010) Knockdown of COPA, identified by loss-of-function screen, induces apoptosis and suppresses tumor growth in mesothelioma mouse model. Genomics 95:210–216. doi:10.1016/j.ygeno.2010.02.002
Smith PM, Elson JL, Greaves LC et al (2014) The role of the mitochondrial ribosome in human disease: searching for mutations in 12S mitochondrial rRNA with high disruptive potential. Hum Mol Genet 23:949–967. doi:10.1093/hmg/ddt490
Bauer TM, Patel MR, Infante JR (2015) Targeting PI3 kinase in cancer. Pharmacol Ther 146:53–60. doi:10.1016/j.pharmthera.2014.09.006
Kim JH, Chu SC, Gramlich JL et al (2005) Activation of the PI3K/mTOR pathway by BCR-ABL contributes to increased production of reactive oxygen species. Blood 105:1717–1723
Hakim S, Bertucci MC, Conduit SE et al (2012) Inositol polyphosphate phosphatases in human disease. Curr Top Microbiol Immunol 362:247–314. doi:10.1007/978-94-007-5025-8_12
Yoon S, Woo SU, Kang JH et al (2012) NF-kappaB and STAT3 cooperatively induce IL6 in starved cancer cells. Oncogene 31:3467–3481. doi:10.1038/onc.2011.517
Cadby G, Mukherjee S, Musk AW et al (2013) A genome-wide association study for malignant mesothelioma risk. Lung Cancer 82:1–8. doi:10.1016/j.lungcan.2013.04.018
Wu C, Hu Z, He Z et al (2011) Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet 43:679–684. doi:10.1038/ng.849
Dogan S, Shen R, Ang DC et al (2012) Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 18:6169–6177. doi:10.1158/1078-0432.CCR-11-3265
Gleeson FC, Kipp BR, Levy MJ et al (2015) Somatic STK11 and concomitant STK11/KRAS mutational frequency in stage IV lung adenocarcinoma adrenal metastases. J Thorac Oncol 10:531–534. doi:10.1097/JTO.0000000000000391
Varghese AM, Sima CS, Chaft JE et al (2013) Lungs don’t forget: comparison of the KRAS and EGFR mutation profile and survival of collegiate smokers and never smokers with advanced lung cancers. J Thorac Oncol 8:123–125. doi:10.1097/JTO.0b013e31827914ea
Rolfo C, Van Der Steen N, Pauwels P et al (2015) Onartuzumab in lung cancer: the fall of Icarus? Expert Rev Anticancer Ther 15:487–489. doi:10.1586/14737140.2015.1031219
Acknowledgments
We thank Päivi Tuominen, Jaana Kierikki, Helinä Hämäläinen, Sauli Savukoski, Tuula Suitiala, Finnish Institute of Occupational Health, and Milja Tikkanen and Tiina Wirtanen, University of Helsinki, for excellent technical assistance. We are also grateful to Ewen MacDonald for the correction of grammar and style. This work was funded by the Finnish Work Environment Fund (no. 112268 to SK; 112269 to HW; 111100 to KHP), Sigrid Jusélius Foundation, Cancer Society of Finland (11/13/2013 to SK; 11/12/2014 to KHP).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Aija Knuuttila received payment for consultancy from Pfizer, Boehringer-Ingelheim, Roche, BMS and for lectures, including service on speakers bureaus, from Pfizer, Lilly, BMS. All other authors declare that they do not have any conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mäki-Nevala, S., Sarhadi, V.K., Knuuttila, A. et al. Driver Gene and Novel Mutations in Asbestos-Exposed Lung Adenocarcinoma and Malignant Mesothelioma Detected by Exome Sequencing. Lung 194, 125–135 (2016). https://doi.org/10.1007/s00408-015-9814-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00408-015-9814-7