Abstract
Purpose
Cervical cancer (CC) patients usually have poor prognosis. The present study aims to find a DNA methylation signature for predicting survival of CC patients.
Methods
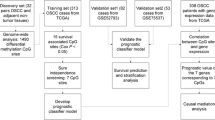
We selected CC patients at pathological stage I–III with corresponding information on radiotherapy and overall survival (OS) from TCGA. Differential expression and methylation analysis was done between patients with and without radiotherapy. We selected feature genes using recursive feature elimination algorithm to build a support vector machine classifier. DNA methylation biomarkers predictive of prognosis were identified using a LASSO Cox-Proportional Hazards model to construct a prognostic scoring model. The classifier and the prognostic model were tested on the training set and the validation set. Nomogram combining risk score and prognostic clinical factors were used.
Results
We obtained 497 differentially expressed genes (DEGs) and 865 differentially methylated genes (DMGs). Fifteen feature genes were selected from the 292 common genes between the DEGs and the DMGs to construct a classification model for radiotherapy. A DNA methylation signature including 10 genes was identified and used to establish a prognostic scoring model. The 10-gene methylation signature could effectively separate patients into two risk groups with markedly different OS time. Predictive capability of the methylation signature was successfully confirmed on the validation set. A nomogram comprised of risk score, radiotherapy, and recurrence was applied, with calibration plots displaying good concordance between predicted and actual OS. The DEGs were involved in 12 KEGG pathways most of which were correlated with metastasis and proliferation of various cancers, such as pathways in cancer, basal cell carcinoma, transcriptional misregulation in cancer and ECM–receptor interaction.
Conclusion
We Identified a 10-gene methylation signature for risk stratification of CC patients at pathological stages I–III, and ten methylation biomarkers might be novel therapeutic targets for CC.






Similar content being viewed by others
References
Mcguire S (2016) World Cancer Report 2014 Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015 Advances in Nutrition 7(2):418
Sakuragi N (2014) Refining insight into cervical cancer progression. Lancet Oncol 15(4):371–372
Baalbergen A, Veenstra Y, Stalpers L (2013) Primary surgery versus primary radiotherapy with or without chemotherapy for early adenocarcinoma of the uterine cervix. Cochrane Database Syst Rev 1(1):0CD06248
Landoni F, Colombo A, Milani R, Placa F, Zanagnolo V, Mangioni C (2017) Randomized study between radical surgery and radiotherapy for the treatment of stage IB–IIA cervical cancer: 20-year update. J Gynecol Oncol 28(3):e34
Kulis M, Esteller M (2010) 2 – DNA methylation and cancer. Adv Genet 70(22):27–56
Issa JPJ (2007) DNA methylation as a therapeutic target in cancer. Clin Cancer Res 13(6):1634–1637
Bhat S, Kabekkodu SP, Noronha A, Satyamoorthy K (2016) Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie 121:298–311
Lorincz AT (2016) Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytol 60(6):501–512
Clarke MA, Luhn P, Gage JC, Bodelon C, Dunn ST, Walker J, Zuna R, Hewitt S, Killian JK, Yan L (2017) Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int J Cancer 141(4):701–710
Zhang J, Yao T, Lin Z, Gao Y (2017) Aberrant methylation of MEG3 functions as a potential plasma-based biomarker for cervical cancer. Sci Rep 7(1):6271. https://doi.org/10.1038/s41598-017-06502-7
Joanna Z, Allison J, Shih-Han L, Esther N, Heidi F, Michal Z, David C, Alexandra S, Salvesen HB, Jacobs IJ (2012) The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet 8(3):e1002517
Yin Z, Sun Y, Ge S, Sun J (2017) Epigenetic activation of WHSC1 functions as an oncogene and is associated with poor prognosis in cervical cancer. Oncol Rep 37(4):2286–2294
Wang Z, Zhang C, Sun L, Liang J, Liu X, Li G, Yao K, Zhang W, Jiang T (2016) FGFR3, as a receptor tyrosine kinase, is associated with differentiated biological functions and improved survival of glioma patients. Oncotarget 51(51):84587
Eberly LE (2003) Correlation and simple linear regression. Radiology 227(3):617–622
Deist TM, Dankers F, Valdes G, Wijsman R, Hsu IC, Oberije C, Lustberg T, van Soest J, Hoebers F, Jochems A, El Naqa I, Wee L, Morin O, Raleigh DR, Bots W, Kaanders JH, Belderbos J, Kwint M, Solberg T, Monshouwer R, Bussink J, Dekker A, Lambin P (2018) Machine learning algorithms for outcome prediction in (chemo)radiotherapy: an empirical comparison of classifiers. Med Phys 45(7):3449–3459. https://doi.org/10.1002/mp.12967
Scholkopf B, Smola AJ (2002) Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT Press, Cambridge
Gerds TA, Kattan MW, Schumacher M, Yu C (2013) Estimating a time-dependent concordance index for survival prediction models with covariate dependent censoring. Stat Med 32(13):2173–2184. https://doi.org/10.1002/sim.5681
Rufibach K (2010) Use of Brier score to assess binary predictions. J Clin Epidemiol 63(8):1250–1260
Robin X (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform 12(1):1–8
Anderson WI, Schlafer DH, Vesely KR (1989) Thyroid follicular carcinoma with pulmonary metastases in a beaver (Castor canadensis). J Wildl Dis 25(4):599–600. https://doi.org/10.7589/0090-3558-25.4.599
Chang Y, An H, Xu L, Zhu Y, Yang Y, Lin Z, Xu J (2015) Systemic inflammation score predicts postoperative prognosis of patients with clear-cell renal cell carcinoma. Br J Cancer 113(4):626–633
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5):E359–E386
Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K, Hou J, Zhang H, Yi S (2017) DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci USA 114(28):7414–7419
Sharareh SR, Kouros O, Chen J, Scarbrough PM, Craciunescu OI, Horton JK, Dressman HK, Blackwell KL, Dewhirst MW (2015) Genomic profiling in locally advanced and inflammatory breast cancer and its link to DCE-MRI and overall survival. Int J Hyperth 31(4):1–10
Ding Y, Yang DZ, Zhai YN, Xue K, Xu F, Gu XY, Wang SM (2017) Microarray expression profiling of long non-coding RNAs in epithelial ovarian cancer. Oncol Lett 14(2):2523–2530
Jiang X, Tan J, Li J, Kivimae S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, Cheyette BN, Yu Q (2008) DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13(6):529–541. https://doi.org/10.1016/j.ccr.2008.04.019
Zhao H, Yang L, Han Y, Li H, Ling Z, Wang Y, Wang E, Wu G (2017) Dact3 inhibits the malignant phenotype of non-small cell lung cancer through downregulation of c-Myb. Int J Clin Exp Pathol 10(12):11580–11587
Al GH, Salgado-Bernabe M, Ck VV, Salcedo-Vargas M, Andrade-Manzano A, Avila-Moreno F, Pina-Sanchez P (2015) Increased expression of HOXB2 and HOXB13 proteins is associated with HPV infection and cervical cancer progression. Asian Pac J Cancer Prev Apjcp 16(4):1349–1353
Yuan H, Kajiyama H, Ito S, Chen D, Shibata K, Hamaguchi M, Kikkawa F, Senga T (2015) HOXB13 and ALX4 induce SLUG expression for the promotion of EMT and cell invasion in ovarian cancer cells. Oncotarget 6(15):13359–13370
Kim H, Hwang JS, Lee B, Hong J, Lee S (2014) Newly identified cancer-associated role of human neuronal growth regulator 1 (NEGR1). J Cancer 5(7):598–608. https://doi.org/10.7150/jca.8052
Kim H, Chun Y, Che L, Kim J, Lee S, Lee S (2017) The new obesity-associated protein, neuronal growth regulator 1 (NEGR1), is implicated in Niemann-Pick disease Type C (NPC2)-mediated cholesterol trafficking. Biochem Biophys Res Commun 482(4):1367–1374
Bedi D, Vig K, Waduwawara S, Samant RS, Yates C (2017) Gene signature for predicting worse relapse-free survival with basal-like breast cancer. Res Rep 1:e1–e112
Agthoven TV, Veldscholte J, Smid M, Agthoven TLAV, Vreede L, Broertjes M, Vries ID, Jong DD, Sarwari R, Dorssers LCJ (2009) Functional identification of genes causing estrogen independence of human breast cancer cells. Breast Cancer Res Treat 114(1):23–30
van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, Look MP, Smid M, Veldscholte J, Sleijfer S, Foekens JA, Dorssers LC (2009) Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol 27(4):542–549. https://doi.org/10.1200/jco.2008.17.1462
Zofia HR, GaLle P, Frédéric C, Céline F, Pauline L, Philippe T, Jean-Michel C, Alain A (2010) YAP1 and VGLL3, encoding two cofactors of TEAD transcription factors, are amplified and overexpressed in a subset of soft tissue sarcomas. Genes Chromosomes Cancer 49(12):1161–1171
Karen G, Quinn MCJ, Wojnarowicz PM, Arcand SL, Manon DL, Véronique B, Jean-Sébastien R, Killary AM, Davis EC, Josée L (2013) VGLL3 expression is associated with a tumor suppressor phenotype in epithelial ovarian cancer. Mol Oncol 7(3):513–530
Ruth E, Walter P, Martina D, Lisa S, Laura W, Martin S, Brigitte W, Markus S, Fabian SD, Judith S (2015) Potential of DNA methylation in rectal cancer as diagnostic and prognostic biomarkers. Br J Cancer 113(7):1035–1045
Atsushi K, Yasuo I, Yoshihiro K, Masakazu S, Koji K, Naoki Y, Naoto T, Jumpei H, Hiroshi A, Tsutomu K (2012) Gene expression signature-based prognostic risk score in patients with primary central nervous system lymphoma. Clin Cancer Res 18(20):5672–5681
Funding
This study was supported by Natural Science Foundation of Shanghai Sicence and Technology Committee, China (No.13ZR1408600), National Natural Science Foundation Youth Project (No. 81702543), Science Foundation of Naval Military Medical University (No. 2017JS05) and Science Foundation of Changhai Hospital (No. 2018JS004).
Author information
Authors and Affiliations
Contributions
SYC, XMY and ZYG participated in the design of this study, and they both performed the statistical analysis. QQY and BWW carried out the study and collected important background information. JZS and RG drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, S., Yu, X., Gu, Z. et al. A 10-gene prognostic methylation signature for stage I–III cervical cancer. Arch Gynecol Obstet 301, 1275–1287 (2020). https://doi.org/10.1007/s00404-020-05524-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-020-05524-3