In an elegant study, Kleinbongard et al. [7] developed a strategy for the quantification of protein expression and phosphorylation in multiple tissue samples taken from various experiments on different gels/membranes by Western blot. Here, they analyze “survival signaling” proteins and their phosphorylation in left ventricular biopsies taken from the previous pig studies on ischemic preconditioning (IPC), ischemic postconditioning (POCO), and remote ischemic conditioning (RIPC). They show that neither the expression nor the phosphorylation of the classical protective signaling proteins (AKT, ERK1/2, GSK-3beta, p38 MAPK, or PKG) were associated with reduced infarct size by different conditioning strategies, and that only STAT3 phosphorylation (when normalized to total STAT3 expression) was consistently associated with cardioprotection obtained by ischemic preconditioning but not by the other conditioning strategies. In the light of the lack of successful translation of many preclinical cardioprotection data to clinical therapy, these results might not be surprising [4]. However, given the abundance of preclinical papers (including studies in pigs) claiming that above survival protein kinases are important mediators of ischemic conditioning, one may question the reproducibility of individual preclinical studies/data and ask a more important question, how to move forward in finding valid molecular targets for cardioprotection (see for a recent position paper [3]).
What do we learn from the above study by Kleinbongard et al. [7]?
-
1.
It is, indeed, important to validate the published experimental animal data by meta-analyses (as done routinely with clinical data) to assess/validate the robustness of an analyzed signaling cascade. Such a meta-analysis could include repetitive data from a single laboratory (as presented by Kleinbongard et al. [7]) or—even better—from different laboratories publishing on the same signaling cascade. For the latter, however, standardized experimental protocols are a pre-requisite. Rigorous quality control of studies across laboratories is challenging; therefore, a recent global European action is aiming for it (http://www.cost.eu/COST_Actions/ca/CA16225).
-
2.
It is also important to decide on the best method to assess changes in the efficacy/activity of a signaling cascade under pathophysiological conditions. Direct measurement of protein activity is often substituted by assessment of its posttranslational modifications (e.g., phosphorylation in absolute terms or normalized to protein expression) or relative expression, the latter might be the weakest indicator for protein activity. The unknown time course of protein activation/inactivation and the possibility to assess only a few time points are further limitations in in vivo experimental studies. As in the study of Kleinbongard et al. [7], pigs might be best suited for such analysis as compared to other species, since repetitive tissue sampling is possible; however, here, the contribution of irreversibly injured and viable tissue to the overall signal is not possible to be quantified in small tissue biopsies. Finally, species dependence of signaling cascades is always a matter of concern (e.g., cardioprotection by postconditioning was not possible in STAT3 knockout mice [1], but STAT3 is possibly not robustly important for postconditioning’s protection in pigs [5]), and the importance of certain proteins for cardioprotection has finally to be proven in humans [6].
-
3.
Problems of reproducibility of data may also be due to biased selection of targets based on a hypothesis that may lead to lack of proper randomization and biased interpretation of the data. Therefore, target findings may also be done using unbiased “fishing” approaches, such as “multi-omics” assays including genomics, epigenomics, transcriptomics, proteomics, and metabolomics, followed by dynamic network analysis to predict the most relevant targets [9]. In acute cardioprotection, epigenomic alterations may be relevant, but so far, there is no study in this field. Technical limitations in full proteomics from heart tissue (see, e.g., [2]) and metabolomics show that these technologies may not be ready for fishing approaches. Therefore, currently, transcriptomics seems to be useful for unbiased target identification at the transcript level (see for reviews [9, 10] and a recent original paper in pigs [8]). Maybe, an easier way of unbiased omics approach is to detect miRNA fingerprints (miRNAs are of limited number and they regulate gene expression) of non-ischemic and ischemia/reperfused hearts with and without conditioning, and identify cardioprotective miRNAs termed “protectomiRs” by a systematic comparison of miRNA expression changes between groups as described [11]. Based on the identified protectomiRs, prediction of their common mRNA targets by bioinformatics methods based on topological or network dynamical approaches can be done (currently, user friendly software is under development, e.g., mirnatarget.com). Nevertheless, all unbiased omics approaches and their bioinformatic evaluation need to be followed by rigorous experimental validation of the predicted targets at the transcript and protein levels (Fig. 1).
Fig. 1 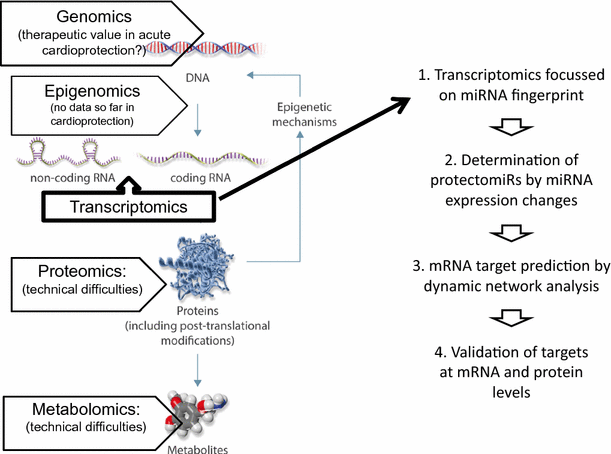
(background image modified from [9])
Proposed way to apply unbiased approach for target identification of cardioprotection based on the determination of miRNA fingerprints in four easy steps
In conclusion: Kleinbongard et al. [7] highlight the importance of validation of individual experimental data by subsequent meta-analyses as done routinely in the clinical setting also for preclinical studies. However, their results also demonstrate that many questions regarding proper target selection (biased vs. unbiased approaches and species dependence) and analysis (protein expression, modification, and activity) need to be solved to facilitate a better translation of experimental findings into daily clinical routine.
References
Boengler K, Buechert A, Heinen Y, Roeskes C, Hilfiker-Kleiner D, Heusch G, Schulz R (2008) Cardioprotection by ischemic postconditioning is lost in aged and STAT3-deficient mice. Circ Res 102:131–135. https://doi.org/10.1161/CIRCRESAHA.107.164699
Gedik N, Kruger M, Thielmann M, Kottenberg E, Skyschally A, Frey UH, Cario E, Peters J, Jakob H, Heusch G, Kleinbongard P (2017) Proteomics/phosphoproteomics of left ventricular biopsies from patients with surgical coronary revascularization and pigs with coronary occlusion/reperfusion: remote ischemic preconditioning. Sci Rep 7:7629. https://doi.org/10.1038/s41598-017-07883-5
Hausenloy DJ, Garcia-Dorado D, Botker HE, Davidson SM, Downey J, Engel FB, Jennings R, Lecour S, Leor J, Madonna R, Ovize M, Perrino C, Prunier F, Schulz R, Sluijter JPG, Van Laake LW, Vinten-Johansen J, Yellon DM, Ytrehus K, Heusch G, Ferdinandy P (2017) Novel targets and future strategies for acute cardioprotection: position paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc Res 113:564–585. https://doi.org/10.1093/cvr/cvx049
Heusch G (2017) Critical issues for the translation of cardioprotection. Circ Res 120:1477–1486. https://doi.org/10.1161/CIRCRESAHA.117.310820
Heusch G, Musiolik J, Gedik N, Skyschally A (2011) Mitochondrial STAT3 activation and cardioprotection by ischemic postconditioning in pigs with regional myocardial ischemia/reperfusion. Circ Res 109:1302–1308. https://doi.org/10.1161/CIRCRESAHA.111.255604
Heusch G, Musiolik J, Kottenberg E, Peters J, Jakob H, Thielmann M (2012) STAT5 activation and cardioprotection by remote ischemic preconditioning in humans: short communication. Circ Res 110:111–115. https://doi.org/10.1161/CIRCRESAHA.111.259556
Kleinbongard P, Skyschally A, Gent A, Pesch M, Heusch G (2017) STAT3 as a common signal of ischemic conditioning—a lesson on “rigor and reproducibility” in preclinical studies on cardioprotection. Basic Res Cardiol. https://doi.org/10.1007/s00395-017-0660-z
Pavo N, Lukovic D, Zlabinger K, Zimba A, Lorant D, Goliasch G, Winkler J, Pils D, Auer K, Jan Ankersmit H, Giricz Z, Baranyai T, Sarkozy M, Jakab A, Garamvolgyi R, Emmert MY, Hoerstrup SP, Hausenloy DJ, Ferdinandy P, Maurer G, Gyongyosi M (2017) Sequential activation of different pathway networks in ischemia-affected and non-affected myocardium, inducing intrinsic remote conditioning to prevent left ventricular remodeling. Sci Rep 7:43958. https://doi.org/10.1038/srep43958
Perrino C, Barabasi A-L, Condorelli G, Davidson SM, De Windt L, Dimmeler S, Engel FB, Hausenloy DJ, Hill JA, Van Laake LW, Lecour S, Leor J, Madonna R, Mayr M, Prunier F, Sluijter JPG, Schulz R, Thum T, Ytrehus K, Ferdinandy P (2017) Epigenomic and transcriptomic approaches in the post-genomic era: path to novel targets for diagnosis and therapy of the ischaemic heart? Position paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc Res 113:725–736. https://doi.org/10.1093/cvr/cvx070
Varga ZV, Giricz Z, Bencsik P, Madonna R, Gyongyosi M, Schulz R, Mayr M, Thum T, Puskas LG, Ferdinandy P (2015) Functional genomics of cardioprotection by ischemic conditioning and the influence of comorbid conditions: implications in target identification. Curr Drug Targets 16:904–911. https://doi.org/10.2174/1389450116666150427154203
Varga ZV, Zvara A, Farago N, Kocsis GF, Pipicz M, Gaspar R, Bencsik P, Gorbe A, Csonka C, Puskas LG, Thum T, Csont T, Ferdinandy P (2014) MicroRNAs associated with ischemia-reperfusion injury and cardioprotection by ischemic pre- and postconditioning: protectomiRs. Am J Physiol Heart Circ Physiol 307:H216–H227. https://doi.org/10.1152/ajpheart.00812.2013
Acknowledgements
PF is the vice chair and RS is a working group leader of the European Cooperation in Science and Technology (COST action CA16225, EU-Cardioprotection). PF holds Grants from the Hungarian National Research, Development, and Innovation Office (OTKA K 109737, OTKA ANN 107803, NVKP 16-1-2016-0017, and VEKOP-2.3.2-16-2016-00002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
PF is the founder and CEO of Pharmahungary Group, a group of R&D companies that develops cardioprotective technologies including miRNA therapeutics.
Rights and permissions
About this article
Cite this article
Schulz, R., Ágg, B. & Ferdinandy, P. Survival pathways in cardiac conditioning: individual data vs. meta-analyses. What do we learn?. Basic Res Cardiol 113, 4 (2018). https://doi.org/10.1007/s00395-017-0661-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00395-017-0661-y


