Abstract
Purpose
Red meat consumption is positively associated with type 1 (T1D) and type 2 (T2D) diabetes. We investigated if red meat consumption increases the risk of latent autoimmune diabetes in adults (LADA) and T2D, and potential interaction with family history of diabetes (FHD), HLA and TCF7L2 genotypes.
Methods
Analyses were based on Swedish case–control data comprising incident cases of LADA (n = 465) and T2D (n = 1528) with matched, population-based controls (n = 1789; n = 1553 in genetic analyses). Multivariable-adjusted ORs in relation to self-reported processed and unprocessed red meat intake were estimated by conditional logistic regression models. Attributable proportion (AP) due to interaction was used to assess departure from additivity of effects.
Results
Consumption of processed red meat was associated with increased risk of LADA (per one servings/day OR 1.27, 95% CI 1.07–1.52), whereas no association was observed for unprocessed red meat. For T2D, there was no association with red meat intake once BMI was taken into account. The combination of high (> 0.3 servings/day vs. less) processed red meat intake and high-risk HLA-DQB1 and -DRB1 genotypes yielded OR 8.05 (95% CI 4.86–13.34) for LADA, with indications of significant interaction (AP 0.53, 95% CI 0.32–0.73). Results were similar for the combination of FHD-T1D and processed red meat. No interaction between processed red meat intake and FHD-T2D or risk variants of TCF7L2 was seen in relation to LADA or T2D.
Conclusion
Consumption of processed but not unprocessed red meat may increase the risk of LADA, especially in individuals with FHD-T1D or high-risk HLA genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Consumption of red meat, especially processed meat, has been associated with an increased risk of type 2 diabetes (T2D) [1] as well as childhood type 1 diabetes (T1D) [2,3,4]. Several compounds in red meat, some of which are particularly abundant in processed meat products, may affect diabetes risk including advanced glycation endproducts (AGEs), sodium, iron, and nitrates, nitrites, and nitrosamines. Proposed mechanisms involve promotion of insulin resistance (e.g., inflammation and oxidative stress) as well as detrimental effects on pancreatic beta cells [5]. The effect may be stronger in genetically susceptible individuals. In support hereof, a study nested within the Health Professionals Follow-up Study reported interaction between red meat and a T2D genetic risk score (GRS) composed of ten single nucleotide polymorphisms (SNPs) in relation to the risk of T2D [6]. Moreover, the association with red meat consumption and islet autoimmunity and T1D has primarily been observed in children carrying high-risk human leukocyte antigen (HLA) genotypes [3, 7].
Latent autoimmune diabetes in adults (LADA) is a common, hybrid form of diabetes with a pathogenesis involving autoimmune destruction of pancreatic beta cells as well as insulin resistance [8]. Genetically, LADA resembles T1D with excess risk conferred by family history of diabetes (FHD) of T1D and HLA genotypes [9, 10]. Genetic similarities with T2D have also been seen including an association with transcription factor 7-like 2 (TCF7L2) [11] and FHD of T2D [9]. Red meat intake may affect the risk of LADA through the same mechanisms potentially linking it to risk of T1D or T2D and the association may depend on genetic factors, but this remains to be explored.
Our objective was to investigate the association between consumption of unprocessed and processed red meat and risk of LADA and T2D, and the potential interaction with FHD of T1D, T2D, and genotypes of HLA and TCF7L2. Furthermore, we aim to explore whether the associations may be mediated by effects of insulin resistance, beta cell function, or autoimmunity.
Subjects and methods
Study design and population
Analyses were based on data from the Swedish population-based case–control study ESTRID (Epidemiological Study of Risk Factors for LADA and Type 2 Diabetes; https://ki.se/imm/estrid; Supplementary Fig. 1) [12]. Incident cases of LADA and T2D are recruited from ANDIS (All New Diabetics in Scania); an extensive registry and biobank aiming to genetically and clinically characterize all new diabetes patients in Scania [13], and ANDiU (All New Diabetics in Uppsala); a sister study in Uppsala County. Since 2010, all incident cases of LADA and a random sample of T2D cases recorded in ANDIS and ANDiU (starting 2012; 5% of ESTRID cases) have been invited to participate by responding to a detailed health- and lifestyle questionnaire. Median time between diagnosis of LADA or T2D and questionnaire response is 6.2 months and 5.0 months, respectively. Population-based, diabetes-free controls are identified through the Swedish Population Register and matched to cases by time and region (incidence-density sampling [14]). Response rate is 80% among cases and 62% among controls. The ESTRID controls provide questionnaire data but no blood samples. For analyses of genetic risk factors, data for diabetes-free controls from the EIRA Study (Epidemiological Investigation on Rheumatoid Arthritis) is used [15]. These controls, from here on referred to as “genetic controls”, are randomly selected from the population and matched to the diabetes cases by age and sex. They answer a similar questionnaire as the cases and provide blood samples for genetic analyses.
Eligible for the present study were all cases and controls included in ESTRID between 2010 and July 2017 with complete covariate information and reported energy intakes within 3 standard deviations (SD) from the loge-transformed sex-specific mean. The analytic sample included 465 LADA cases, 1528 T2D cases, and 1789 controls. The genetic controls were collected 2005–2014, aged ≥ 35 years, and with complete covariate information including ≥ 1 of the genetic factors (HLA or/and TCF7L2) (n = 1553). The study was approved by the Regional Ethical Review Board in Stockholm and all participants provided informed consent.
Classification of diabetes
Classification into diabetes subtype was based on age at diagnosis, glutamic acid decarboxylase autoantibodies (GADA), and fasting C-peptide. Details of the serological assay methods have been described previously [13]. GADA was determined using enzyme-linked immunosorbent assay (ELISA) with 84% sensitivity and 98% specificity at 10.7 IU/mL cut-off level [16]. Values > 250 IU/mL were censored at 250 IU/mL. C-peptide concentration was measured using Cobas e601 analyzer (Roche Diagnostics, Germany) or IMMULITE 2000 (Siemens Healthcare Diagnostics Product Ldt., UK). Cases of LADA had age ≥ 35 years, GADA ≥ 10 IU/mL, and C-peptide ≥ 0.2 nmol/L (IMMULITE) or ≥ 0.3 nmol/L (Cobas e 601). T2D was defined as age ≥ 35 years, GADA < 10 IU/mL, and C-peptide ≥ 0.60 nmol/L (IMMULITE) or ≥ 0.72 nmol/L (Cobas e 601). Homeostatic model assessment of insulin resistance (HOMA-IR) and beta cell function (HOMA-B) were calculated based on fasting levels of plasma glucose and C-peptide [17].
Diet and other covariate assessment
Dietary intake was assessed using a 132-item semi-quantitative food frequency questionnaire (FFQ). Participants were asked to report how often, on average over the previous year, they had consumed various foods by indicating one of eight pre-defined frequency categories ranging from “0 times per month” to “≥ 3 times per day”. The patients were specifically instructed to report their habitual intake reflecting the year preceding diagnosis. Eleven questions concerned red meat consumption; five items on unprocessed red meat [pork, beef/veal, minced meat, offal (liver/kidney), and other meat], and six on processed red meat [sausages/hot dogs, Falun sausage, other sausages, cold cuts (ham/salami), bacon, black pudding, and liver paté]. Energy intake (kcal/day) was estimated based on age- and sex-specific serving sizes combined with nutrition values from the Swedish National Food Agency database. The FFQ has been validated for nutrients using fourteen 24-h recall interviews, with Spearman correlation coefficients of 0.65 for macronutrients and 0.62 for micronutrients [18]. The genetic controls answered identical questions on red meat consumption. As a whole, their FFQ was somewhat shorter with 124 items, hampering comparability of estimated energy intake.
Information on non-dietary covariates was derived from the questionnaire. Height and bodyweight were used to calculate BMI (kg/m2). Highest attained education level was categorized as primary school, upper secondary school, or university. Four pre-defined response options ranging from sedentary to regularly active were used to assess leisure-time physical activity during the preceding year (prior to diagnosis for patients). Smoking habits were categorized into never, former, or current smoker. Average daily alcohol intake was estimated from the FFQ and categorized into none, 0.01–4.9 g/day, 5–14.9 g/day, and ≥ 15 g/day. FHD of T1D (FHD-T1D) was defined as a first-degree relative with diabetes onset age < 40 years combined with insulin therapy, otherwise defined as FHD of T2D (FHD-T2D). When used in confounder adjustment in the main analyses, FHD was dichotomized into none or ≥ 1 relative with diabetes. The corresponding information was available for the genetic controls, with the exceptions of energy intake and FHD.
Genotyping
Genotyping of patients was based on blood samples analyzed at the Clinical Research Center in Malmö, Sweden, using iPlex Gold Technology (Sequenom, San Diego, CA, USA). For a subset, missing genotypes were imputed using Infinium CoreExome v1.1 (Illumina, USA), based on the Haplotype Reference Consortium (https://www.haplotype-reference-consortium.org/; version r1.1 2016) reference panel. For genetic controls, genotyping was based on GWAS data from Illumina Global Screening array or Infinium Illumina 300K immunochip custom array (Illumina, USA). Based on three SNPs within the HLA region (rs3104413, rs2854275, rs9273363), participants were genotyped according to previously described methodology [19]. Based on the literature [20, 21] and frequency distributions in our study population, participants were categorized as carriers of high-risk HLA genotypes (DR4-DQ8; DR4/3-DQ8; DR3/4; DR3/3; DR4/4, and DRB1*0301-DQA1*0501-DQB1*0201) or low-/intermediate-risk genotypes (DR3/x; DR4/x; DR4-DQ7, and DRx/x, where x = neither DR4 nor DR3). For TCF7L2 rs7903146, participants were classified as risk genotype carriers if they had at ≥ 1 risk allele (i.e., TT or TC).
Statistical analysis
Characteristics for patients and controls were presented as proportions, means, or medians (skewed data), with SD (means) or interquartile range (IQR; medians). Characteristics of study participants are also presented by quartiles of consumption of unprocessed red meat and processed red meat in supplementary tables. Chi-square test (proportions), Student’s t test (means), and Kruskal–Wallis H (medians) tests were used to calculate two-tailed p values.
Conditional logistic regression models were used to estimate odds ratios (OR) and 95% confidence intervals (CI) of LADA and T2D in relation to genotypes, FHD, and consumption of unprocessed and processed red meat. ESTRID controls were used in all analyses except those including genetic covariates. Model 1 was adjusted for age and sex. In model 2, additional adjustment was made for education, physical activity, smoking, alcohol, energy intake (continuous), and FHD. Model 3 additionally included BMI (continuous), a potential mediator. With regard to red meat consumption, results from model 3 is discussed in the text unless otherwise specified. Model 1 adjustment was used for the association between genotypes and diabetes (corresponding to Supplementary Fig. 2), and the analysis of LADA and T2D in relation to FHD (≥ 1 first degree relative) was adjusted according to model 3 except energy intake but with mutual adjustment for FHD-T2D and FHD-T1D. All covariates included were chosen based on previous knowledge on our study population and the literature. Red meat consumption was analyzed in quartiles (based on distribution among controls) and continuously per one daily serving increment [median serving size was 107.4 g (interquartile range, IQR 55.9) for unprocessed red meat and 46.3 g (IQR 26.1) for processed red meat]. The lowest consumption category was used as reference. Restricted cubic splines (model 3) with three knots were used to explore potential nonlinear relationships. Linear regression models (model 3) were used to explore the change in GADA (Tobit regression to account for the right-censoring at 250 IU/mL), HOMA-IR, and HOMA-B (loge-transformed) per one daily serving increment in red meat intake.
Attributable proportion (AP) due to interaction was estimated to examine the presence of interaction, defined as departure from additivity of effects, between processed red meat consumption [low (lowest quartile; 0–0.3 servings/day) or high (upper three quartiles)] and genotypes of HLA and TCF7L2 as well as FHD-T1D and FHD-T2D on the risk of LADA and T2D. These analyses were adjusted according to model 3, except that genetic analyses did not include energy intake and FHD. AP > 0 with a 95% CI not including 0 indicate significant positive interaction [22].
Sensitivity analyses (model 3) were performed by additional adjustments for diet (fruits, vegetables, fatty fish, snacks, coffee, sweetened beverages) and mutual adjustment for unprocessed and processed meat. We also made restrictions to patients responding to the questionnaire within 6 months of diagnosis, currently on diabetes treatment that includes ‘diet modification’, and not reporting ‘major lifestyle changes’ after diagnosis of diabetes.
Statistical Analysis Software 9.4 (SAS Institute, USA) and Stata Statistical Software 14.2 (StataCorp, USA) (for spline modeling) were used for statistical analyses.
Results
Characteristics
In comparison with T2D, patients with LADA were less overweight and obese, less insulin resistant, but had worse beta cell function (Table 1). The proportion of high-risk HLA genotype carriers was equal in individuals with T2D and controls, but considerably higher in LADA. Individuals in the highest quartiles of red meat intake also had higher energy intake compared to those in the lowest quartiles (Supplementary Table 1). Among those in the highest quartile of unprocessed red meat intake, 40.7% were also in the highest quartile of processed red meat intake (not shown in table).
Red meat intake, LADA and T2D
Processed red meat consumption was positively associated with LADA; each additional daily serving was associated with OR 1.27 (95% CI 1.07–1.52) and an OR of 1.53 (1.08–2.16) was observed in the highest vs. lowest quartile. No association was seen with unprocessed red meat (OR per daily serving 0.69, 95% CI 0.46–1.03). For LADA, restricted cubic spline analysis indicated an increased risk that appeared to be linear for processed red meat consumption exceeding 1.5 servings/day, but no indication of an association with unprocessed red meat (Fig. 1).
Restricted cubic spline models estimating ORs (solid line) with 95% CIs (dashed line) of LADA and type 2 diabetes in relation to consumption of unprocessed red meat and processed red meat, adjusted according to Model 3. The distribution of red meat intake in the study population is presented in the histogram at the bottom of each panel
T2D was not associated with processed or unprocessed red meat consumption after adjustment for BMI (Table 2, Fig. 1).
Genotypes, FHD, red meat, LADA and T2D
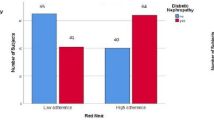
As shown previously in this population [9, 23], LADA was strongly associated with HLA genotypes and to a lesser extent also with genotypes of TCF7L2 (Supplementary Fig. 2). Similarly, LADA was more strongly associated with FHD-T1D than with FHD-T2D. The combination of high-risk HLA genotypes and high consumption of processed red meat conferred an OR of 8.05 (95% CI 4.46–13.34) for LADA with AP estimated at 0.53 (95% CI 0.32–0.73) (Fig. 2). High consumption of processed red meat combined with FHD-T1D indicated similar results (OR 6.61, 95% CI 3.79–11.51; with AP 0.44, 95% CI − 0.14 to 1.02) (Table 3). There was no evidence of interaction between processed red meat intake and TCF7L2 genotype or FHD-T2D in relation to LADA.
T2D was associated with TCF7L2 risk genotypes and FHD-T2D, but not with high-risk HLA genotypes and FHD-T1D (Supplementary Fig. 2). However, no evidence of interaction was found between intake of red meat and genotypes of TCF7L2 or FHD-T2D on the risk of T2D (Table 3).
Red meat intake, HOMA, and GADA
In T2D individuals, one additional daily serving of processed red meat was inversely associated with HOMA-B (− 6.7%, p = 0.0056) and positively associated with HOMA-IR (8.1%, p = 0.0029) (Supplementary Table 2). Unprocessed red meat intake was inversely associated with HOMA-B (− 12.2%, p = 0.0096) but not with HOMA-IR (Supplementary Table 2). In LADA patients, a decrease in HOMA-B and increase in HOMA-IR in relation to red meat intake, particularly processed red meat, was suggested but not significantly so (Supplementary Table 2). No associations were observed between red meat intake and GADA.
Sensitivity analysis
Additional adjustment for dietary factors had minor impact on the associations (OR per 1 daily serving of processed meat was 1.26, 95% CI 1.06–1.51 for LADA; and 1.09, 95% CI 0.94–1.25, for T2D). Neither did mutual adjustment for unprocessed and processed meat alter the observed associations between red meat intake and LADA or T2D. The increased risk of LADA per one daily serving increment in processed meat intake was observed also when restricting the analysis to patients responding to the questionnaire within 6 months of diabetes diagnosis (OR 1.21, 95% CI 0.95–1.55). Similarly, the results did not change materially neither when excluding patients reporting ‘diet modification’ as current diabetes treatment (OR per additional serving was 1.21, 95% CI 0.99–1.47, for LADA), nor when excluding patients reporting ‘major lifestyle changes’ after diagnosis (LADA n = 156, OR 1.24, 95% CI 1.01–1.52).
Discussion
These novel findings indicate that consumption of processed red meat increases the risk of LADA, especially in combination with high-risk HLA genotypes or FHD-T1D. In contrast, for T2D no associations with processed or unprocessed red meat were observed, and neither TCF7L2 genotype nor FHD-T2D influenced these findings. Processed red meat consumption was, however, associated with increased insulin resistance and worse beta cell function among individuals with T2D, with similar but non-significant associations for LADA.
Our findings are in line with previous observations in T1D in children; Recently, Syrjälä and colleagues demonstrated a positive association between total meat intake and islet autoimmunity and childhood T1D [4]. Furthermore, in a case–control study, an increased risk of T1D was found in relation to total meat intake [2]. Maternal intake of red meat and meat products during lactation [3], but not during pregnancy [24,25,26], has been positively associated with islet autoimmunity or/and T1D. Moreover, biomarkers of red meat fat [7], but not the meat intake [27] were associated with autoimmunity in genetically susceptible children.
An increased risk of LADA observed for intake of processed, but not unprocessed, red meat may suggest that underlying mechanisms are related to compounds specific to, or found at higher levels in, processed meat products. Furthermore, the highest risk of LADA in relation to processed red meat intake was observed when combined with high-risk HLA genotype or FHD-T1D. Indeed, many of the compounds hypothesized to be responsible for a positive association with both autoimmune diabetes and T2D are more abundant in processed red meat with suggested mechanisms involving adverse effects on beta cell function. This may be a potential explanation for our finding; individuals at high genetic risk may already have compromised beta cell function and hence be more susceptible to the adverse effects of high processed meat intake. The lack of interaction between processed red meat intake and genotypes of TCF7L2 or FHD-T2D on the risk of LADA, and the overall lack of association regarding T2D, support the potential involvement of autoimmune-related genetic susceptibility. Compounds that potentially link red meat intake to diabetes risk include AGEs, which are proinflammatory compounds formed in the preparation and cooking processes of red meat, particularly processed meat. In rodent studies, high AGE diets have been demonstrated to induce insulin secretory dysfunction, and a low AGE diet decreased the incidence of autoimmune diabetes in nonobese diabetic mice and improved insulin resistance and beta cell function in mice with T2D [28]. Other compounds of interest in processed meat products are nitrates and nitrites, which are added as preservatives and potentially converted to nitrosamines [5]. Toxic effect of nitrosamines on beta cells has been supported by studies in rodents [29] and dietary nitrites and nitrosamines have been positively associated with childhood T1D in case–control studies [30, 31]. Potential adverse effects have also been ascribed sodium, which is another micronutrient found at high levels in processed meat and has been associated with T2D risk and insulin resistance [32].
Several prospective studies have reported that consumption of red meat, particularly processed red meat, is associated with increased risk of T2D [33]. We found a similar positive association, but this was fully attenuated after adjustment for BMI. This may suggest that the association is mediated through effects on BMI. In fact, BMI has previously been suggested to partly explain the associations between red meat intake and T2D [34,35,36], and red meat intake has been positively associated with weight gain [37]. However, contrary to our findings, in these studies the associations remain, although attenuated, also after adiposity adjustment. One possible explanation may be that longitudinal studies often adjust for historical BMI (assessed at baseline or other prior diagnosis timepoint), whereas we adjust for BMI at diagnosis (index time for controls) which may yield more complete adjustment. The fact that we observed increased insulin resistance and worse beta cell function in relation to red meat consumption for T2D, with similar indications also for LADA, speak in favour of direct adverse effects. There were no indications that consumption of processed red meat affect LADA risk by triggering autoimmunity. In this context it is worth noting that a recent publication based on prospective data found no association between red meat consumption and risk of rheumatoid arthritis [38]. An association with insulin resistance is supported by a recent longitudinal study reporting positive associations, also after adiposity adjustment, for HOMA-IR in relation to total animal protein and protein from meat [39]. We did not observe any interaction between processed red meat intake and genotypes or FHD in relation to T2D. This contrasts a previous U.S. study which found an interaction with a T2D-GRS including TCF7L2 (rs12255372) [6] but is in line with a Swedish study using a different GRS also including TCF7L2 (rs7903146) [40].
Strengths of the present study include the population-based design, extensive information on diet and essential confounding factors, and the large number of incident cases of LADA. Given that LADA is relatively rare, a case–control design is an efficient way to achieve enough cases for viable analyses [14]. However, since exposure data is collected retrospectively, recall bias is a concern. Importantly, dietary data was collected in close conjunction with diagnosis and participants were instructed to report their habits preceding diagnosis. Furthermore, the observed associations remained in sensitivity analysis restricted to patients with the shortest duration of diabetes at study inclusion. In addition, our results for T2D before adjustment for BMI are in line with previous, prospective studies [34, 36]. Another concern is that patients may have made dietary modifications following diagnosis and reported their modified dietary regimen. However, this would lead to overestimations of the observed positive associations only if patients had increased their intakes of red meat after diagnosis. This seems unlikely; the national guidelines for diabetes patients recommend increased intake of unsaturated fats (at the expense of saturated fats) and the general dietary guidelines recommend limiting the intake of red meat, particularly processed meat, as part of a healthy diet. Of note, the observed ORs were largely unchanged in sensitivity analyses excluding patients reporting being on ‘diet modification’ treatments or having had major lifestyle changes after diagnosis. The use of external genetic controls and the fact that it was not possible to adjust for energy intake in the interaction analysis with HLA and TCF7L2 is a limitation, but energy intake did not seem to have a great impact on the estimates in the main analysis using the internal controls. The genetic controls had slightly lower processed red meat intake compared to the internal controls (mean: 0.7 serv/day and 0.8 serv/day, respectively), which may potentially lead to overestimated OR associated with processed red meat intake. However, any such impact on the study findings seem limited, since the interaction analyses with genotypes showed similar results as those obtained in interaction analyses, where FHD was used as an indicator of genetic susceptibility, i.e., when the ESTRID controls were used and energy intake was accounted for.
In conclusion, we present novel findings that consumption of processed red meat may be a risk factor for LADA, independent of adiposity, and potentially most detrimental in individuals with HLA-conferred susceptibility or FHD-T1D. These findings add support for a role of processed red meat in the development of autoimmune diabetes in children and adults. This is important considering that established modifiable risk factors for primary prevention of autoimmune diabetes are still lacking.
References
Neuenschwander M, Ballon A, Weber KS, Norat T, Aune D, Schwingshackl L, Schlesinger S (2019) Role of diet in type 2 diabetes incidence: umbrella review of meta-analyses of prospective observational studies. BMJ 366:l2368. https://doi.org/10.1136/bmj.l2368
Muntoni S, Mereu R, Atzori L et al (2013) High meat consumption is associated with type 1 diabetes mellitus in a Sardinian case–control study. Act Diabetol 50:713–719. https://doi.org/10.1007/s00592-012-0385-2
Niinistö S, Takkinen HM, Uusitalo L et al (2015) Maternal intake of fatty acids and their food sources during lactation and the risk of preclinical and clinical type 1 diabetes in the offspring. Acta Diabetol 52:763–772. https://doi.org/10.1007/s00592-014-0673-0
Syrjälä E, Nevalainen J, Peltonen J et al (2019) A joint modeling approach for childhood meat, fish and egg consumption and the risk of advanced islet autoimmunity. Sci Rep 9:7760. https://doi.org/10.1038/s41598-019-44196-1
Wolk A (2017) Potential health hazards of eating red meat. J Intern Med 281:106–122. https://doi.org/10.1111/joim.12543
Qi L, Cornelis MC, Zhang C, van Dam RM, Hu FB (2009) Genetic predisposition, Western dietary pattern, and the risk of type 2 diabetes in men. Am J Clin Nutr 89:1453–1458. https://doi.org/10.3945/ajcn.2008.27249
Virtanen SM, Niinistö S, Nevalainen J et al (2010) Serum fatty acids and risk of advanced beta-cell autoimmunity: a nested case–control study among children with HLA-conferred susceptibility to type I diabetes. Eur J Clin Nutr 64:792–799. https://doi.org/10.1038/ejcn.2010.75
Tuomi T, Santoro N, Caprio S, Cai M, Weng J, Groop L (2014) The many faces of diabetes: a disease with increasing heterogeneity. Lancet 383:1084–1094. https://doi.org/10.1016/S0140-6736(13)62219-9
Hjort R, Alfredsson L, Andersson T et al (2017) Family history of type 1 and type 2 diabetes and risk of latent autoimmune diabetes in adults (LADA). Diabetes Metab 43:536–542. https://doi.org/10.1016/j.diabet.2017.05.010
Mishra R, Chesi A, Cousminer DL et al (2017) Relative contribution of type 1 and type 2 diabetes loci to the genetic etiology of adult-onset, non-insulin-requiring autoimmune diabetes. BMC Med 15:88. https://doi.org/10.1186/s12916-017-0846-0
Andersen MK, Hansen T (2019) Genetic aspects of latent autoimmune diabetes in adults: a mini-review. Curr Diabetes Rev 15:194–198. https://doi.org/10.2174/1573399814666180730123226
Rasouli B, Andersson T, Carlsson PO, Grill V, Groop L, Martinell M, Storm P, Tuomi T, Carlsson S (2016) Smoking and the risk of LADA: results from a Swedish population-based case-control study. Diabetes Care 39:794–800. https://doi.org/10.2337/dc15-2348
Ahlqvist E, Storm P, Käräjämäki A et al (2018) Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6:361–369. https://doi.org/10.1016/S2213-8587(18)30051-2
Vandenbroucke JP, Pearce N (2012) Case–control studies: basic concepts. Int J Epidemiol 41:1480–1489. https://doi.org/10.1093/ije/dys147
Stolt P, Bengtsson C, Nordmark B, Lindblad S, Lundberg I, Klareskog L, Alfredsson L, EIRA study group (2003) Quantification of the influence of cigarette smoking on rheumatoid arthritis: results from a population based case–control study, using incident cases. Ann Rheum Dis 62:835–841. https://doi.org/10.1136/ard.62.9.835
Rahmati K, Lernmark A, Becker C, Foltyn-Zadura A, Larsson K, Ivarsson SA, Törn C (2008) A comparison of serum and EDTA plasma in the measurement of glutamic acid decarboxylase autoantibodies (GADA) and autoantibodies to islet antigen-2 (IA-2A) using the RSR radioimmunoassay (RIA) and enzyme linked immunosorbent assay (ELISA) kits. Clin Lab 54:227–235
Oxford Centre for Diabetes, Endocrinology & Metabolism. Diabetes Trial Unit. HOMA Calculator. https://www.dtu.ox.ac.uk/homacalculator. Accessed 2 June 2013
Messerer M, Johansson SE, Wolk A (2004) The validity of questionnaire-based micronutrient intake estimates is increased by including dietary supplement use in Swedish men. J Nutr 134:1800–1805. https://doi.org/10.1093/jn/134.7.1800
Nguyen C, Varney MD, Harrison LC, Morahan G (2013) Definition of high-risk type 1 diabetes HLA-DR and HLA-DQ types using only three single nucleotide polymorphisms. Diabetes 62:2135–2140. https://doi.org/10.2337/db12-1398
Hagopian WA, Sanjeevi CB, Kockum I, Landin-Olsson M, Karlsen AE, Sundkvist G, Dahlquist G, Palmer J, Lernmark A (1995) Glutamate decarboxylase-, insulin-, and islet cell-antibodies and HLA typing to detect diabetes in a general population-based study of Swedish children. J Clin Investig 95:1505–1511. https://doi.org/10.1172/JCI117822
Törn C, Gupta M, Nikitina Zake L, Sanjeevi CB, Landin-Olsson M (2003) Heterozygosity for MICA5.0/MICA5.1 and HLA-DR3-DQ2/DR4-DQ8 are independent genetic risk factors for latent autoimmune diabetes in adults. Hum Immunol 64:902–909. https://doi.org/10.1016/s0198-8859(03)00158-7
Andersson T, Alfredsson L, Källberg H, Zdravkovic S, Ahlbom A (2005) Calculating measures of biological interaction. Eur J Epidemiol 20:575–579. https://doi.org/10.1007/s10654-005-7835-x
Hjort R, Löfvenborg JE, Ahlqvist E et al (2019) Interaction between overweight and genotypes of HLA, TCF7L2, and FTO in relation to the risk of latent autoimmune diabetes in adults and type 2 diabetes. J Clin Endocrinol Metab 104:4815–4826. https://doi.org/10.1210/jc.2019-00183
Lamb MM, Myers MA, Barriga K, Zimmet PZ, Rewers M, Norris JM (2008) Maternal diet during pregnancy and islet autoimmunity in offspring. Pediatr Diabetes 9:135–141. https://doi.org/10.1111/j.1399-5448.2007.00311.x
Brekke HK, Ludvigsson J (2010) Daily vegetable intake during pregnancy negatively associated to islet autoimmunity in the offspring—the ABIS study. Pediatr Diabetes 11:244–250. https://doi.org/10.1111/j.1399-5448.2009.00563.x
Virtanen SM, Uusitalo L, Kenward MG et al (2011) Maternal food consumption during pregnancy and risk of advanced β-cell autoimmunity in the offspring. Pediatr Diabetes 12:95–99. https://doi.org/10.1111/j.1399-5448.2010.00668.x
Virtanen SM, Nevalainen J, Kronberg-Kippilä C et al (2012) Food consumption and advanced β cell autoimmunity in young children with HLA-conferred susceptibility to type 1 diabetes: a nested case–control design. Am J Clin Nutr 95:471–478. https://doi.org/10.3945/ajcn.111.018879
Coughlan MT, Yap FY, Tong DC et al (2011) Advanced glycation end products are direct modulators of β-cell function. Diabetes 60:2523–2532. https://doi.org/10.2337/db10-1033
de la Monte SM, Tong M, Lawton M, Longato L (2009) Nitrosamine exposure exacerbates high fat diet-mediated type 2 diabetes mellitus, non-alcoholic steatohepatitis, and neurodegeneration with cognitive impairment. Mol Neurodegener 4:54. https://doi.org/10.1186/1750-1326-4-54
Dahlqvist GG, Blom LG, Persson LA, Sandström AI, Wall SG (1990) Dietary factors and the risk of developing insulin dependent diabetes in childhood. BMJ 300:1302–1306. https://doi.org/10.1136/bmj.300.6735.1302
Virtanen SM, Jaakkola L, Räsänen L, Ylönen K, Aro A, Lounamaa R, Akerblom HK, Tuomilehto J, Childhood Diabetes in Finland Study Group (1994) Nitrate and nitrite intake and the risk for type 1 diabetes in Finnish children. Diabet Med 11:656–662. https://doi.org/10.1111/j.1464-5491.1994.tb00328.x
Muris DM, Houben AJ, Schram MT, Stehouwer CD (2012) Microvascular dysfunction is associated with a higher incidence of type 2 diabetes mellitus: a systematic review and meta-analysis. Arterioscler Thromb Vasc Biol 32:3082–3094. https://doi.org/10.1161/ATVBAHA.112.300291
Schwingshackl L, Hoffmann G, Lampousi AM, Knüppel S, Iqbal K, Schwedhelm C, Bechthold A, Schlesinger S, Boeing H (2017) Food groups and risk of type 2 diabetes mellitus: a systematic review and meta-analysis of prospective studies. Eur J Epidemiol 32:363–375. https://doi.org/10.1007/s10654-017-0246-y
Pan A, Sun Q, Bernstein AM, Schulze MB, Manson JE, Willett WC, Hu FB (2011) Red meat consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. Am J Clin Nutr 94:1088–1096. https://doi.org/10.3945/ajcn.111.018978
Ericson U, Sonestedt E, Gullberg B, Hellstrand S, Hindy G, Wirfält E, Orho-Melander M (2013) High intakes of protein and processed meat associate with increased incidence of type 2 diabetes. Br J Nutr 109:1143–1153. https://doi.org/10.1017/S0007114512003017
InterAct Consortium, Bendinelli B, Palli D et al (2013) Association between dietary meat consumption and incident type 2 diabetes: the EPIC-InterAct study. Diabetologia 56:47–59. https://doi.org/10.1007/s00125-012-2718-7
Mozaffarian D, Hao T, Rimm EB, Willett WC, Hu FB (2011) Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med 364:2392–2404. https://doi.org/10.1056/NEJMoa1014296
Sundström B, Ljung L, Di Giuseppe D (2019) Consumption of meat and dairy products is not associated with the risk for rheumatoid arthritis among women: a population-based cohort study. Nutrients 11:2825. https://doi.org/10.3390/nu11112825
Chen Z, Franco OH, Lamballais S, Ikram MA, Schoufour JD, Muka T, Voortman T (2020) Associations of specific dietary protein with longitudinal insulin resistance, prediabetes and type 2 diabetes: the Rotterdam Study. Clin Nutr 39:242–249. https://doi.org/10.1016/j.clnu.2019.01.021
Ericson U, Hindy G, Drake I, Schulz CA, Brunkwall L, Hellstrand S, Almgren P, Orho-Melander M (2018) Dietary and genetic risk scores and incidence of type 2 diabetes. Genes Nutr 13:13. https://doi.org/10.1186/s12263-018-0599-1
Acknowledgements
Open access funding provided by Karolinska Institute.
Funding
ESTRID was supported the Swedish Research Council (GA 2018-03035), the Swedish Research Council for Health, Working life and Welfare (GA 2018-00337), the Swedish Diabetes Foundation, and the Novo Nordisk Foundation. EIRA received funding from the Swedish Research Council, the Swedish Research Council for Health, Working Life and Welfare, the Swedish Rheumatic Foundation, the AFA Insurance Company, and Stockholm County Council. Funding for ANDIS was provided by the Swedish Research Council and the European Research Council Advanced Researcher grant (GA 269045) to LG and ALF—the Swedish Research Council funding for clinical research.
Author information
Authors and Affiliations
Contributions
All authors contributed to the interpretation of results, and critically reviewed and revised and approved the final version of the manuscript. Contributions to the data collection was made by SC, JEL, and TA (ESTRID), LG, EA, and TT (ANDIS), and LA (EIRA). TA contributed statistical expertise. SC contributed to the conception and design of the study and thoroughly reviewed and revised the manuscript. JEL contributed to the conception and design of the study, analyzed the data, drafted and revised the manuscript, and is the guarantor of this work. As such, JEL had full access to the data and takes responsibility for the integrity of the data, and the accuracy of the data analysis and the study report.
Corresponding author
Ethics declarations
Conflict of interest
The Authors report no conflicts of interest relevant to this article.
Ethical approval
The study was approved by the Regional Ethical Review Board in Stockholm and all participants provided informed consent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Löfvenborg, J.E., Ahlqvist, E., Alfredsson, L. et al. Consumption of red meat, genetic susceptibility, and risk of LADA and type 2 diabetes. Eur J Nutr 60, 769–779 (2021). https://doi.org/10.1007/s00394-020-02285-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-020-02285-2